What is the structure of green fluorescent protein GFP?
structure summary. The green fluorescent protein (GFP) is a protein composed of 238 amino acid residues (26.9 kDa) that exhibits bright green fluorescence when exposed to light in the blue to ultraviolet range.
What is the wavelength of GFP light?
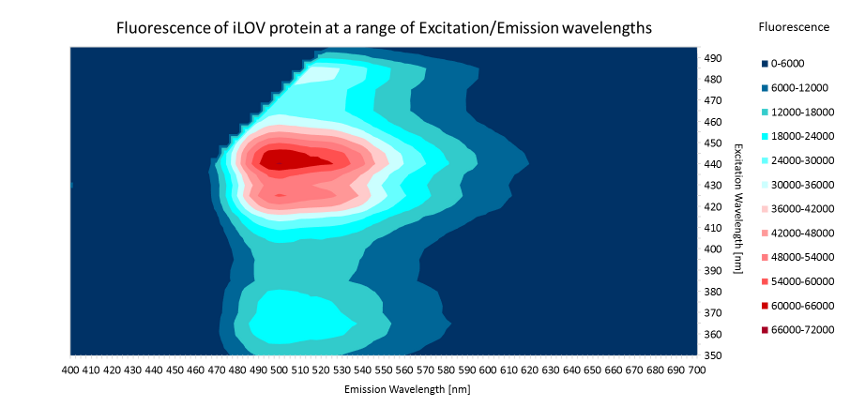
GFP is excited at a wavelength of 488nm and has an emission peak at ~507nm. However, high-resolution crystal structures of GFP allow scientists to manipulate its protein structure and engineer color variants that emit at different wavelengths 3, 4 .
What is the ICAO code for green fluorescent protein?
For the airport with that ICAO airport code, see Pembrey Airport. The green fluorescent protein (GFP) is a protein that exhibits bright green fluorescence when exposed to light in the blue to ultraviolet range. The label GFP traditionally refers to the protein first isolated from the jellyfish Aequorea victoria and is sometimes called avGFP.
What is the fluorescence quantum yield of GFP?
Its emission peak is at 509 nm, which is in the lower green portion of the visible spectrum. The fluorescence quantum yield (QY) of GFP is 0.79. The GFP from the sea pansy (Renilla reniformis) has a single major excitation peak at 498 nm.
See more

What light does GFP fluoresce under?
Green fluorescent protein (GFP) is a protein in the jellyfish Aequorea Victoria that exhibits green fluorescence when exposed to light. The protein has 238 amino acids, three of them (Numbers 65 to 67) form a structure that emits visible green fluorescent light.
At what wavelengths did GFP show absorbance peaks?
The absorption spectrum of the green fluorescent protein is special in the sense that it exhibits two maxima, one at 479 nm, which is ascribed to the excitation of a deprotonated (anion) chromophore, and a main peak at 397 nm, which supposedly correlates to a protonated (neutral) chromophore (3).
How does GFP promote fluorescence?
1. GFP is a barrel shape with the fluorescent portion (the chromophore) made up of just three amino acids. When this chromophore absorbs blue light, it emits green fluorescence.
What is the wavelength of EGFP?
As demonstrated in Figure 1, the red-shifted variants, typified by EGFP, have a single excitation peak centered at about 488 nm, with an emission peak wavelength of 509 nm.
How long does GFP fluorescence last?
The half-life of unmodified GFP is approximately 26 h;8 thus, it takes several days for the passively transferred protein to degrade leading to an overestimation of transduction achieved at early time points.
What are the excitation and emission wavelengths for GFP S65T?
This GFP variant had the longest-wavelength excitation maximum yet described, 504 nm, as compared with 490 nm for S65T (Table 1).
Why does GFP glow under UV light?
Scientists knew that GFP glows because three of its amino acids form a fluorophore, a chemical group that absorbs and emits light.
Why is it that GFP fluorescent green?
Solutions of purified GFP look yellow under typical room lights, but when taken outdoors in sunlight, they glow with a bright green color. The protein absorbs ultraviolet light from the sunlight, and then emits it as lower-energy green light.
Can you see GFP with naked eye?
The transformants showing high expression of the gfp gene had the normal mycelia pigmentation altered, displaying a bright green-yellowish color, visible with the naked eye on the plates, without the aid of any kind of fluorescent light or special filter set.
What wavelength of light excites GFP?
GFP can be excited by the 488 nm laser line and is optimally detected at 510 nm.
How do I check my GFP fluorescence?
Flow cytometry and fluorescent microscopy are two conventional tools to detect the GFP signal; flow cytometry is an effective and sensitive technique to quantitatively analyze fluorescent intensity, while fluorescent microscopy can visualize the subcellular location and expression of GFP.
What is the difference between GFP and EGFP?
The main difference between GFP and EGFP is that the GFP (stands for Green Fluorescent Protein) is a protein that exhibits bright green fluorescence when exposed to blue light whereas the EGFP (stands for Enhanced Green Fluorescence Protein) exhibits stronger fluorescence than GFP.
What is the excitation and emission maxima of EGFP?
EGFP is a fluorescent compound with an excitation peak at 489 nm and an emission peak at 511 nm. It can be excited using a 488 nm laser paired with a 530/30 nm bandpass filter, a configuration that can be found, for example, in the BD FACSAria™ II.
How large is GFP RFP in kDa?
RFP is approximately 25.9 kDa. The excitation maximum is 558 nm, and the emission maximum is 583 nm. The first fluorescent protein to be discovered, green fluorescent protein (GFP), has been adapted to identify and develop fluorescent markers in other colors.
What wavelength is mCherry?
mCherry is a fluorescent compound with an excitation peak at 587 nm and an emission peak at 610 nm.
What is the shape of a GFP protein?
The protein structure, first reported in 1996, is an eleven β-sheet-containing “barrel” shape , with the chromophore concealed at the center of the structure, shielded from quenching by aqueous solvent. This tightly-packed structure explains the importance of the entire GFP protein, which is almost completely required to maintain fluorescent ...
Why Green Fluorescent Protein?
It has a fluorescent emission wavelength in the green portion of the visible spectrum (hence the name), which is due to a chromophore formed from a maturation reaction of three specific amino acids at the center of the protein (Ser65, Tyr66, and Gly67). When first discovered, one of the most surprising aspects of GFP was the fact that the chromophore forms spontaneously and without additional co-factors, substrates, or enzymatic activity – it only requires the presence of oxygen during maturation. This meant that the protein could be taken directly from A. Victoria and expressed in any organism while still maintaining fluorescence.
What is a GFP biosensor?
Biosensors: A wide array of GFP-based fluorescent biosensors has been designed to detect a variety of intracellular conditions, including ion (such as Ca2+) concentrations and pH, using a range of strategies such as FRET, calmodulin, and others. Review Addgene's collection of fluorescent biosensors here.
What is FACS in flow cytometry?
Fluorescence-activated cell sorting (FACS): This is a type of flow cytometry that separates mixtures of cells into distinct populations based on fluorescent signal. Thus, FACS can be used to separate cells expressing GFP from cells that are not.
Why do plasmids have GFP?
Cell marking/selection: Expression constructs like plasmids often include GFP as a marker to help identify which cells have successfully taken up the plasmid. This can serve as an alternative to antibiotic selection. Plasmids of this type may have the GFP under the control of an additional promoter from that of the gene of interest, or expressed from the same transcript as the gene of interest, but after an internal ribosome entry site (IRES). This is oftentimes used in conjuction with FACS (see below).
What is FRET in biology?
Förster resonance energy transfer (FRET): This is used to study the interactions between two proteins or between two domains of a protein that undergoes conformational change. Typcially two fluorescent proteins with overlapping excitation/emission spectra are used; one fused to each protein or domain being tested. Find FRET plasmids here.
Why is GFP used in cell fate studies?
Developmental/transgenic uses: Because of its stability, GFP can be used in lineage tracking capacities in cell fate studies. It can also be used, when put under control of promoters of interest, to visualize the developmental stage at which these promoters are active.
What is green fluorescent protein?
Green fluorescent proteins are being used for more and more applications in molecular and cellular biology. As a result of the variety of applications several variants form the original wild type green fluorescent protein (wtGFP) have been developed. Several of these variants have different excitation and emission spectra than wtGFP.
What are the mutations in FL600?
In terms of filter sets for the FL600 microplate fluorescence reader, the most commonly used mutations to the wtGFP protein can be categorized into three groups: (1) the red-shifted variants; (2) the wild type like variants; and (3) the blue emitting variants. As demonstrated in Figure 1, the red-shifted variants, typified by EGFP, have a single excitation peak centered at about 488 nm, with an emission peak wavelength of 509 nm. The wild type like variants have their primary excitation peak centered on 395 nm, with an emission peak at 509 nm while the blue emitting mutants generally have an excitation peak at around 380 nm and an emission peak near 460 nm (Figure 1).
What filter does the FL600 use?
The red-shifted mutants generally only require the standard "fluorescein" filter set of the 485/20 excitation and 530/25 emission, both of which are standard filters on the FL600. Likewise, the blue emitting mutants use the 360/40 excitation and 460/40 emission filters, also standard on the FL600. A.
What is green fluorescent protein?
Green Fluorescent Protein (GFP) is a versatile biological marker for monitoring physiological processes, visualizing protein localization, and detecting transgenic expression in vivo. GFP can be excited by the 488 nm laser line and is optimally detected at 510 nm.
What is the color of the GFP in 3T3 cells?
NIH 3T3 cells that were transiently transfected with a Green Fluorescent Protein (GFP) expression vector, then plated and allowed to attach and proliferate. The cells were fixed and labeled with our Invitrogen Alexa Fluor 594 conjugate of the anti-GFP antibody. Cells expressing GFP show dual labeling of both GFP (green fluorescence) and the anti-GFP antibody (red fluorescence). In this overlay of fluorescence and differential interference contrast (DIC) micrographs, the GFP-transfected cells exhibit green and red signals that overlap to yield yellow, and DAPI stains the nuclei with a light-blue fluorescence. In the cells that are not transfected, the DAPI-stained nuclei exhibit a bright blue fluorescence.
What is anti-GFP antibody?
Anti-GFP antibodies provide a convenient method for visualizing GFP, especially when amplification of the fluorescent protein of interest is necessary to overcome a dim or degraded signal. Our anti-GFP antibodies are easily incorporated into standard immunostaining protocols for cell and tissue analysis.
What is GFP in microscopy?
GFP is the most popular, most widely used genetically encoded fluorescent probe. Several factors contribute to the popularity of GFP including (i) ...
How big is GFP?
GFP is big. GFP is a 28 kDa protein that resembles a cylinder with a length of 4.2 nm and a diameter of about 2.4 nm ( Hink et al., 2000 ). The complete beta-barrel is necessary for its fluorescence and therefore GFP cannot be downsized by deleting residues.
Why is GFP so popular?
Several factors contribute to the popularity of GFP including (i) fast and complete maturation to functional, fluorescent protein in almost all organisms and cell types, (ii) no need to add a co-factor, (iii) easy visualization with standard filter sets on a fluorescence microscope, and finally (iv) good toleration in fusion proteins.
How to avoid autofluorescence?
Another solution to avoid autofluorescence is offered by bioluminescent proteins. These probes generate light by a chemical reaction and hence, there is no need to excite the sample. Although bioluminescent probes are usually dim relative to fluorescent probes, recent engineering efforts have increased the brightness of these genetically encoded light emitters (i.e. Takai, 2015 and Iwano et al., 2018 ).
How are GFP fusion proteins made?
GFP fusion proteins are made by connecting its DNA code to the cDNA encoding for another protein. Effectively, it equips the protein of interest with an additional protein module that can report on location. This approach enables the study of proteins in cells, but not other biomolecules of interest e.g. DNA, RNA, and lipids. Still, these molecules can be indirectly visualized by using protein domains that specifically bind the molecule of interest. For instance, pleckstrin homology domains can be used to detect specific phosphoinositides ( Varnai and Balla, 2008) ( Figure 2).
Which fluorescent protein is better for detection?
Therefore, the use of bright red fluorescent proteins (e.g. mScarlet) or infrared fluorescent proteins ( Chernov et al., 2017) may improve detection. Since longer wavelengths have better tissue penetration, (infra)red fluorescent proteins are also a better choice for thicker samples.
When N- and C-terminal fusions are not tolerated, the alternative is to insert the GFP?
When N- and C-terminal fusions are not tolerated the alternative is to insert the GFP into the coding sequence of the protein of interest. One of the reasons that this works well is that the N- and C-terminus of GFP itself are relatively close, thereby minimizing the disruption of the target protein.
What is the structure of fluorescent protein?
Although this simple amino acid motif is commonly found throughout nature, it does not generally result in fluorescence. What is unique to the fluorescent protein is that the location of this peptide triplet resides in the center of a remarkably stable barrel structure consisting of 11 beta -sheets folded into a tube.
How many nanometers does a green fluorescent protein absorb?
A protonated form, the predominant state, has an excitation maximum at 395 nanometers, and a less prevalent, unprotonated form that absorbs at approximately 475 nanometers.
How to adapt fluorescent proteins?
In order to adapt fluorescent proteins for use in mammalian systems, several basic modification s of the wild-type green fluorescent protein were undertaken and are now found in all commonly used variants. The first step was to optimize the maturation of fluorescence to a 37-degree Celsius environment. Maturation of the wild-type fluorophore is quite efficient at 28 degrees, but increasing the temperature to 37 degrees substantially reduces overall maturation and results in decreased fluorescence. Mutation of the phenylalanine residue at position 64 ( Phe64) to leucine results in improved maturation of fluorescence at 37 degrees, which is at least equivalent to that observed at 28 degrees. This mutation is present in the most popular varieties of fluorescent proteins derived from Aequorea victoria, but is not the only mutation that improves folding at 37 degrees as other variants have been discovered.
How does denaturation affect fluorescent proteins?
Denaturation of green fluorescent protein destroys fluorescence, as might be expected, and mutations to residues surrounding the tripeptide fluorophore can dramatically alter the fluorescence properties. The packing of amino acid residues inside the beta barrel is extremely stable, which results in a very high fluorescence quantum yield (up to 80 percent). This tight protein structure also confers resistance to fluorescence variations due to fluctuations in pH, temperature, and denaturants such as urea. The high level of stability is generally altered in a negative manner by mutations in green fluorescent protein that perturb fluorescence, resulting in a reduction of quantum yield and greater environmental sensitivity. Although several of these defects can be overcome by additional mutations, derivative fluorescent proteins are generally more sensitive to the environment than the native species. These limitations should be seriously considered when designing experiments with genetic variants.
What are two examples of multiple fluorescent protein labeling in living cells?
The opossum kidney cortex proximal tubule epithelial cell ( OK line) presented in Figure 1 (a) was transfected with a cocktail of fluorescent protein variants fused to peptide signals that mediate transport to either the nucleus (enhanced cyan fluorescent protein; ECFP ), the mitochondria (DsRed fluorescent protein; DsRed2FP ), or the microtubule network (enhanced green fluorescent protein; EGFP ). A similar specimen consisting of human cervical adenocarcinoma epithelial cells ( HeLa line) is depicted in Figure 1 (b). The HeLa cells were co-transfected with sub-cellular localization vectors fused to cyan ( mTurquoise) and yellow ( mVenus) fluorescent protein coding sequences (Golgi complex and the nucleus, respectively), as well as the "Fruit" protein, mCherry, targeting the mitochondrial network.
Why are fluorescent probes important?
Thus, the development of far red fluorescent probes would be extremely useful for the examination of thick specimens and entire animals. Given the success of fluorescent proteins as reporters in transgenic systems, the use of far red fluorescent proteins in whole organisms will become increasingly important in the coming years.
Why are fluorescent proteins used in live cells?
In live cells, fluorescent proteins are most commonly employed to track the localization and dynamics of proteins, organelles, and other cellular compartments.
What is superfolder GFP?
Superfolder GFP is a basic (constitutively fluorescent) green fluorescent protein published in 2005, derived from Aequorea victoria. It is reported to be a very rapidly-maturing weak dimer. Oligomerization. Organism.
Does Superfolder GFP have excerpts?
No excerpts have been added for Superfolder GFP. Excerpts are snippets from publications that capture key information about this protein that does not easily fit into one of the existing fields (such as a summary, motivation, or observation). Add an excerpt.
