
How many chromatids per chromosome exist following anaphase 2?
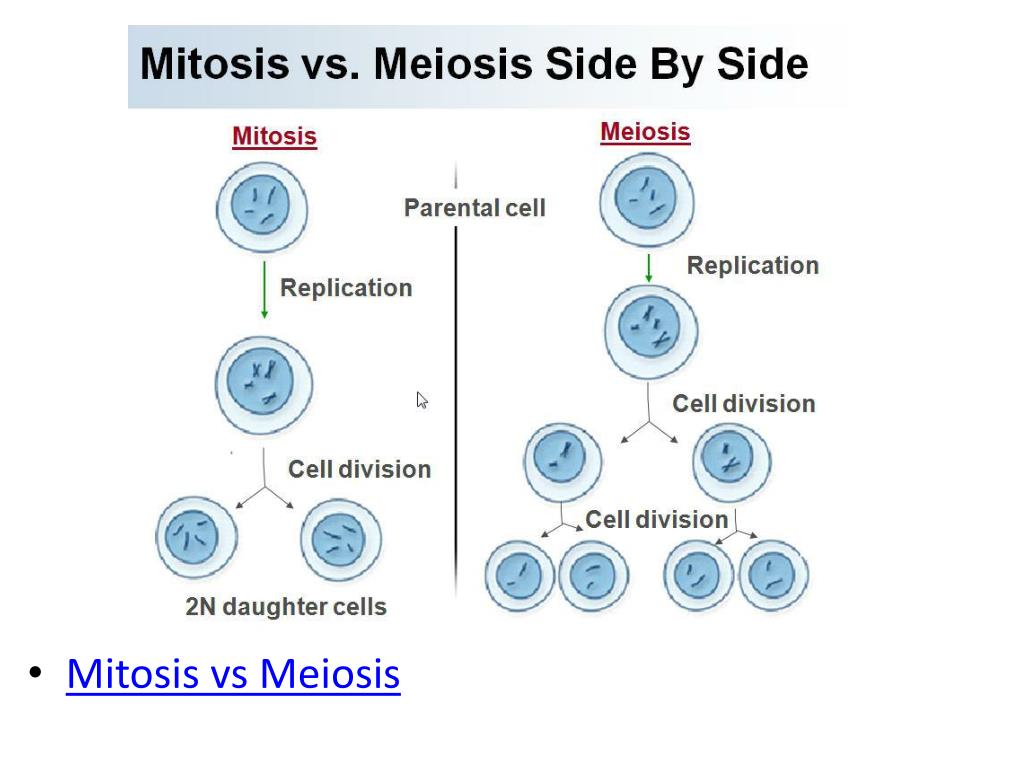
During anaphase, we now have a total of 16 chromosomes and 16 chromatids – in short, each chromatid is now a chromosome. Similarly, in humans, there are 92 chromosomes present and 92 chromatids during anaphase. These numbers remain the same during telophase.
How many centromeres are visible in anaphase?
There are 46 chromosomes in a human cell, and hence, 46 centromeres (or equal to the number of chromosomes in a cell i.e. 2n) at Anaphase I; whereas, in Anaphase II there are 46 (2n) centromeres and 46 (2n) chromatids. Where are the centromeres of homologous chromosomes located?
How many centromeres are visible during prophase?
are present only during prophase. consist entirely of DNA. are present only during mitosis. ... one origin of replication, two telomeres, and many centromeres. one telomere, two centromeres, and many origins of replication. ... They are clearly visible as defined bodies under the light microscope.
When do centromeres divide?
The centromeres divide at the beginning of anaphase so that the two chromatids of each chromosome become separated from each other and are now considered to be chromosomes. The sister chromosomes move toward opposite poles of the cell with their centromeres in the lead. Likewise, what happens to centromeres during cell division?
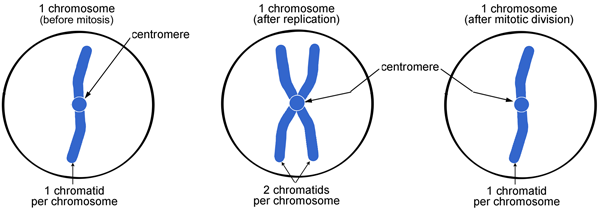
Does one chromosome have centromere?
The monocentric chromosome is a chromosome that has only one centromere in a chromosome and forms a narrow constriction. Monocentric centromeres are the most common structure on highly repetitive DNA in plants and animals.
Does a single chromatid have a centromere?
A chromatid is a replicated chromosome having two daughter strands joined by a single centromere (the two strands separate during cell division to become individual chromosomes).
Do chromosomes have two centromeres?
In general, each chromosome contains a single region of centromeric DNA where the centromere and kinetochore are assembled. However, genome rearrangements can lead to fusion of two different chromosomes, often resulting in a dicentric chromosome on which two centromeres are physically linked.
How many centromere is present in a chromosome?
Note: A chromosome contains 2 chromatids and a centromere.
Do chromosomes always have 2 chromatids?
Chromosomes can have 1 or 2 chromatids, depending on whether they have replicated.
What is the difference between single and double-stranded chromosomes?
A single chromosome contains only on single long unbranched double-stranded DNA molecule that displays the typical double-helix structure. This double-strands DNA is formed by one phosphate group alternating with one desoxyribose group coupled to each other by nucleic acids (adenine, guanine, thymine en cytosine).
What happens if a chromosome has two centromeres?
A dicentric chromosome is an abnormal chromosome with two centromeres. It is formed through the fusion of two chromosome segments, each with a centromere, resulting in the loss of acentric fragments (lacking a centromere) and the formation of dicentric fragments.
Where is the centromere on a chromosome?
The centromere is a very specific part of the chromosome. When you look at the chromosomes, there's a part that is not always right in the middle, but it's somewhere between one-third and two-thirds of the way down the chromosome. It's called the centromere.
What would happen without centromeres?
Without the centromere, no kinetochore would form and cells could not segregate their chromosomes. Thus, the centromere is of crucial importance for chromosome segregation and mitotic control.
How do you calculate the number of centromeres?
1:327:13How to count the Number of Chromosomes and DNA molecules ...YouTubeStart of suggested clipEnd of suggested clipSo once it is duplicated a chromosome will be having two chromatids rule number 2 how to count theMoreSo once it is duplicated a chromosome will be having two chromatids rule number 2 how to count the number of DNA molecule to count the number of DNA molecule. Simply count the number of chrome.
How many centrosomes are in a cell?
two centrosomesThe majority of cells in our body contain either one or two centrosomes, depending on their position within the cell cycle. A G1 cell contains a single centrosome composed of two centrioles.
Do sister chromatids have centromere?
Sister chromatids are two identical copies of the same chromosome formed by DNA replication, attached to each other by a structure called the centromere.
How many chromatids are in a centromere?
Chromatid The two “sister” chromatids are joined at a constricted region of the chromosome called the centromere.
Is a single chromatid a chromosome?
The chromosome consists of a single chromatid and is decondensed (long and string-like). The DNA is copied. The chromosome now consists of two sister chromatids, which are connected by proteins called cohesins.
Does each sister chromatid have a centromere?
A sister chromatid refers to the identical copies (chromatids) formed by the DNA replication of a chromosome, with both copies joined together by a common centromere.
How many chromatids are in a chromosome?
Each chromosome consists of 2 sister chromatids.
What is the centromere of a holocentric chromosome?
Unlike monocentric chromosomes, in holocentric chromosomes the entire length of the chromosome act s as the centromere. In holocentric chromosomes there is not one primary constriction but the centromere has many CenH3 loci spread over the whole chromosome. Examples of this type of centromere can be found scattered throughout the plant and animal kingdoms, with the most well-known example being the nematode Caenorhabditis elegans .
How does centromere misregulation affect cancer?
Notably, overexpression of many centromere genes have been linked to cancer malignant phenotypes. Overexpression of these centromere genes can increase genomic instability in cancers . Elevated genomic instability on one hand relates to malignant phenotypes; on the other hand, it makes the tumor cells more vulnerable to specific adjuvant therapies such as certain chemotherapies and radiotherapy. Instability of centromere repetitive DNA was recently shown in cancer and aging.
What type of DNA is a centromere?
Any piece of DNA with the point centromere DNA sequence on it will typically form a centromere if present in the appropriate species. The best characterised point centromeres are those of the budding yeast, Saccharomyces cerevisiae.
How are dicentric chromosomes formed?
A dicentric chromosome is an abnormal chromosome with two centromeres. It is formed through the fusion of two chromosome segments, each with a centromere, resulting in the loss of acentric fragments (lacking a centromere) and the formation of dicentric fragments. The formation of dicentric chromosomes has been attributed to genetic processes, such as Robertsonian translocation and paracentric inversion. Dicentric chromosomes have important roles in the mitotic stability of chromosomes and the formation of pseudodicentric chromosomes.
What is the centromere in mitosis?
When cells enter mitosis, the sister chromatids (the two copies of each chromosomal DNA molecule resulting from DNA replication in chromatin form) are linked along their length by the action of the cohesin complex. It is now believed that this complex is mostly released from chromosome arms during prophase, so that by the time the chromosomes line up at the mid-plane of the mitotic spindle (also known as the metaphase plate), the last place where they are linked with one another is in the chromatin in and around the centromere.
What is the role of centromeres in cell division?
The physical role of the centromere is to act as the site of assembly of the kinetochores – a highly complex multiprotein structure that is responsible for the actual events of chromosome segregation – i.e. binding microtubules and signalling to the cell cycle machinery when all chromosomes have adopted correct attachments to the spindle, so that it is safe for cell division to proceed to completion and for cells to enter anaphase.
How many chromosomes are neocentromeres found on?
This phenomenon is most well known from human clinical studies and there are currently over 90 known human neocentromeres identified on 20 different chromosomes.
What is the centromere in cytogenetics?
And these P and Q arms are a part of what we use when we do cytogenetics to say how many chromosomes are present in a cell and what chromosome number they are.
What is the name of the region of a chromosome that separates into a short arm and?
Centromere. A centromere is a constricted region of a chromosome that separates it into a short arm (p) and a long arm (q). During cell division, the chromosomes first replicate so that each daughter cell receives a complete set of chromosomes.
Why are chromosomes held together during cell division?
During cell division, this is the place where the chromosomes, when they're undergoing replication, that they're held together so that the chromosomes don't lose their sister chromatid during the cell division process . Julie A. Segre, Ph.D.
Where is the centromere located?
When you look at the chromosomes, there's a part that is not always right in the middle, but it's somewhere between one-third and two-thirds of the way down the chromosome. It's called the centromere. That's the part where the cell's chromosomes are constricted, and they're a little bit tighter, and it almost looks like a little ball in ...
Can you add videos to your watch history?
Videos you watch may be added to the TV's watch history and influence TV recommendations. To avoid this, cancel and sign in to YouTube on your computer.
What is centromere dysfunction?
Centromere dysfunction is also suspected to play a role in cancer cells, which display massive chromosome imbalance of the type that would be expected if the sorting of chromosomes during cell division failed.
What is the role of centromere dysfunction in miscarriage?
Centromere dysfunction leading to problems with chromosome sorting is believed to play a role in many instances of miscarriage, in which inherited centromere disorders may result in early embryonic death. Centromere dysfunction is also suspected to play a role in cancer cells, which display massive chromosome imbalance of the type ...
What is the cytoskeleton at the centromere?
At the centromere, elements of the cell’s cytoskeleton assemble and attach. First, a complex of proteins called the kinetochore assembles around the centromere region of DNA; then, mitotic spindle fibers attach to the kinetochore. The other end of these fibers are anchored to opposite ends of the parent cell, which will shortly split ...
How does DNA work in a cell?
To ensure that a full copy of its DNA is given to each daughter cell, a cell first makes a complete copy of its DNA. The two copies stick together, ultimately condensing to form sister chromatids, until they are pulled apart during cell division. The centromere of the chromosome provides a binding site for the mitotic spindle fiber ...
What is the point on a chromosome where mitotic spindle fibers attach to pull sister?
The centromere is the point on a chromosome where mitotic spindle fibers attach to pull sister chromatids apart during cell division. When a cell seeks to reproduce itself, it must first make a complete copy of each of its chromosomes, to ensure that their daughter cell receives a full complement of the parent cell’s DNA.
What type of centromeres do humans use?
Humans and most eukaryotic cells use regional centromeres. These are centromeres where mitotic spindle binding is determined, not by a precise sequence of DNA, but by a combination of characteristics working together to signal the location of a centromere.
Why are the two sister chromatids together called a single chromosome?
The two sister chromatids combined are often referred to as a single chromosome because they are packaged tightly together – but each contains all the information of the original chromosome, so when they split, each becomes a complete chromosome containing all of the information contained in the parent cell’s original chromosome.
What is the chromatin structure of the centromere region?
The chromatin structure of centromere region is epigenetically unique. Centromere-specific histone H3 is a primary determinant in establishing functional centromeres and kinetochore assembly sites. In humans, the centromere-specific histone H3 variant is CENP-A. It replaces the normal H3 histone at sites where centromeres reside and kinetochores attach chromosomes to spindle fibers. This specialized centromeric chromatin is the foundation for binding of other centromere-associated proteins.
What is the centromere of a chromosome?
Figure 2. Centromeric DNAs vary among different eukaryotes. Centromeres can be defined as small point centromeres, encoded by define d DNA sequences, such as in S. cerevisiae. Other organisms assemble regional centromeres on a larger portion of the chromosome, spanning several hundreds of kilobases. Assembly of regional centromeres is epigenetic or less dependent on DNA sequence. In the nematode C. elegans, the centromere is assembled along the entire length of the chromosome. Common proteins (green) are found at centromeres of diverse organisms, despite fundamental differences in DNA sequence.
How are centromeres organized?
Centromeres vary remarkably in size and sequence across different species and are organized as point, regional, or holocentric ( Figure 2 ). The centromeres of the unicellular Saccharomyces cerevisiae, or baking yeast, are referred to as point centromeres. Sequences spanning approximately 125 base pairs (bp) direct centromere assembly and position. This ∼125 bp sequence encompasses three elements (CDE I, II, and III) that define the centromeres of all 16 budding yeast chromosomes. Over 100 kinetochore proteins form the point centromere on each chromosome, creating an attachment site for a single microtubule. In contrast to the budding yeast point centromere, the centromeres of larger eukaryotes are defined as ‘regional’ centromeres that span more than several kilobases (kb) in size. Regional centromeres are determined epigenetically, that is, without a complete dependence on primary DNA sequence. For instance, the eight centromeres in the pathogenic yeast Candida albicans are 4–5 kb in size, but each differs in primary DNA sequence. Centromeres in the fission yeast Schizosaccharomyces pombe span 40–100 kb, and while they share some of the same repetitive sequences, each has different central core regions. Humans, other primates, plants, Drosophila, and mice all have large regional centromeres composed of repetitive DNA elements. Centromere repeats in humans are made up of alpha-satellite DNA, a ∼171 bp sequence or monomer that is iteratively repeated. Alpha-satellite monomers are arranged head-to-tail and organized into repeated arrays called higher order repeats (HORs) that range from 0.2 to 5 megabases (Mb) and are chromosome specific. Holocentric or holokinetic chromosomes of the microscopic roundworm Caenorhabditis elegans as well as many types of insects do not have a single point or regional centromere. Instead, the kinetochore extends along the entire length of the chromosome, and microtubule attachments span the length of the chromosomes. This centromere type can also be found within other species, such as protozoans, green algae, invertebrates, and plants.
How do centromeres ensure chromosome segregation?
Therefore, to maintain centromeres and ensure accurate chromosome segregation, CENP-A nucleosomes must be inherited across generations through the germline. In this chapter we discuss three aspects of maternal centromere inheritance. First, we propose mechanisms for maintaining CENP-A nucleosomes through the prolonged prophase arrest in mammalian oocytes. Second, we review mechanisms by which selfish centromeres bias their transmission through female meiosis. Third, we discuss regulation of centromere size through early embryonic development.
What are centromeres made of?
Centromere repeats in humans are made up of alpha-satellite DNA, a ∼171 bp sequence or monomer that is iteratively repeated.
How many bp are centromeres?
Budding yeast ( S. cerevisiae) centromeres are 125 bp and are composed of three distinct elements, two of which are conserved (I and III).
Where is the centromere located on a human chromosome?
Human chromosomes 2, 4–12, 17, 18, and X are submetacentric. In acrocentric chromosomes, the centromere is located quite near the distal end of the p-arm. Human chromosomes 13–15, 21, 22, and Y are acrocentric. Telocentric chromosomes have their centromere at the very end of the chromosome.
What is a pars site?
parS is usually called a "centromere-like" site , but is sometimes referred to as a "plasmid centromere”, which is why the quote uses the phrase so-called “plasmid centromeres”.
Do prokaryotes have centromeres?
No, prokaryotes do not have centromeres. They do, however, generally have a somewhat analogous structure used during cell division to partition the replicated chromosomes and plasmids between the two daughter cells. The analogous place on the chromosome or plasmid is the parS element, a DNA sequence on the chromosome or plasmid which is part of the parABS system. This system, consisting of two proteins ( parA and parB) along with parS, forms a mechanism to segregate the copies of a chromosome or plasmid for the daughter cells.
What is the meaning of "back up"?
Making statements based on opinion; back them up with references or personal experience.
Is a chromosome linear or eukaryotic?
Eukaryotic chromosomes are always linear. ... In contrast, prokaryotic chromosomes are either completely devoid of centromeres or carry the so-called “plasmid centromeres” which are not essential (with a few exceptions, such as Caulobacter) (57,–60).
Overview
The centromere links a pair of sister chromatids together during cell division. This constricted region of chromosome connects the sister chromatids, creating a short arm (p) and a long arm (q) on the chromatids. During mitosis, spindle fibers attach to the centromere via the kinetochore.
The physical role of the centromere is to act as the site of assembly of the kine…
Position
In humans, centromere positions define the chromosomal karyotype, in which each chromosome has two arms, p (the shorter of the two) and q (the longer). The short arm 'p' is reportedly named for the French word "petit" meaning 'small'. The position of the centromere relative to any particular linear chromosome is used to classify chromosomes as metacentric, submetacentric, acrocentric, te…
Centromere types
An acentric chromosome is fragment of a chromosome that lacks a centromere. Since centromeres are the attachment point for spindle fibers in cell division, acentric fragments are not evenly distributed to daughter cells during cell division. As a result, a daughter cell will lack the acentric fragment and deleterious consequences could occur.
Chromosome-breaking events can also generate acentric chromosomes or acentric fragments.
Sequence
There are two types of centromeres. In regional centromeres, DNA sequences contribute to but do not define function. Regional centromeres contain large amounts of DNA and are often packaged into heterochromatin. In most eukaryotes, the centromere's DNA sequence consists of large arrays of repetitive DNA (e.g. satellite DNA) where the sequence within individual repeat elements is similar but not identical. In humans, the primary centromeric repeat unit is called α-satellite (or a…
Inheritance
Since centromeric DNA sequence is not the key determinant of centromeric identity in metazoans, it is thought that epigenetic inheritance plays a major role in specifying the centromere. The daughter chromosomes will assemble centromeres in the same place as the parent chromosome, independent of sequence. It has been proposed that histone H3 variant CENP-A (Centromere Protein A) is the epigenetic mark of the centromere. The question arises whether there must be …
Structure
The centromeric DNA is normally in a heterochromatin state, which is essential for the recruitment of the cohesin complex that mediates sister chromatid cohesion after DNA replication as well as coordinating sister chromatid separation during anaphase. In this chromatin, the normal histone H3 is replaced with a centromere-specific variant, CENP-A in humans. The presence of CENP-A is believed to be important for the assembly of the kinetochore on the centromere. CENP-C has be…
Centromeric aberrations
In rare cases, neocentromeres can form at new sites on a chromosome as a result of a repositioning of the centromere. This phenomenon is most well known from human clinical studies and there are currently over 90 known human neocentromeres identified on 20 different chromosomes. The formation of a neocentromere must be coupled with the inactivation of the previous centromere, since chromosomes with two functional centromeres (Dicentric chromoso…
Dysfunction and disease
It has been known that centromere misregulation contributes to mis-segregation of chromosomes, which is strongly related to cancer and miscarriage. Notably, overexpression of many centromere genes have been linked to cancer malignant phenotypes. Overexpression of these centromere genes can increase genomic instability in cancers. Elevated genomic instability on one hand relates to malignant phenotypes; on the other hand, it makes the tumor cells more …