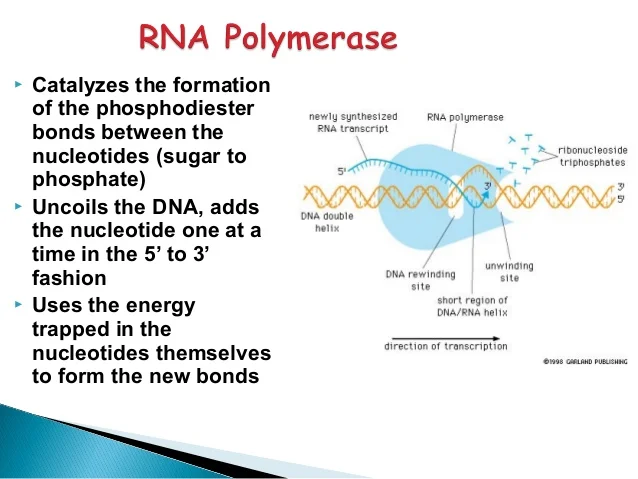
Does DNA and RNA have phosphodiester bonds?
In DNA and RNA, the phosphodiester bond is the linkage between the 3' carbon atom of one sugar molecule and the 5' carbon atom of another, deoxyribose in DNA and ribose in RNA. Strong covalent bonds form between the phosphate group and two 5-carbon ring carbohydrates (pentoses) over two ester bonds.
Do phosphodiester bonds link nucleotides in RNA?
The phosphodiester bond links a 3' carbon to a 5' carbon in DNA and RNA. During the reaction of two of the hydroxyl groups in phosphoric acid with a hydroxyl group in two other molecules two ester bonds in a phosphodiester group are formed.
What types of bonds are in RNA?
In RNA, nucleotides are bonded by a phosphodiester bond. ... Phosphodiester bonds make up the backbone of the strands of DNA. ... Strong covalent bonds form between the phosphate group and two 5-carbon ring carbohydrates (pentoses) over two ester bonds.
What bonds hold RNA together?
Single-stranded RNA can also form many secondary structures in which a single RNA molecule folds over and forms hairpin loops, stabilized by intramolecular hydrogen bonds between complementary bases.
What is the importance of phosphodiester bonds in DNA and RNA molecules?
Phosphodiester bonds are ester bonds that form between sugar and phosphate to form the backbone of nucleic acids. Phosphodiester bond function is crucial to stabilize the structure of DNA and RNA. DNA and RNA are responsible for the inheritance of genetic material and protein expression.
Are phosphodiester bonds formed in transcription?
The phosphodiester bond connects the 3' carbon atom of one sugar molecule to the 5' carbon atom of another, deoxyribose in DNA and ribose in RNA. The "backbones of DNA and RNA" are made up of phosphodiester links. Thus, phosphodiester bonds play an important role in transcription.
What bonds link nucleotides?
Nucleotides are joined together by covalent bonds between the phosphate group of one nucleotide and the third carbon atom of the pentose sugar in the next nucleotide.
Which of these enzymes forms phosphodiester bonds in RNA or DNA select all that apply?
DNA polymerase I, DNA ligase, and Topoisomerase I each catalyze the formation of phosphodiester bonds that are vital (directly or indirectly) to replication of DNA.
What is a phosphodiester bond in biology?
A phosphodiester bond is a covalent bond between phosphate and 2 sugars (hydroxyl groups). A covalent bond involves the sharing of a pair of electr...
Why is a phosphodiester bond important?
A phosphodiester bond is vital for the maintenance of the structural stability of nucleic acids. Nucleic acids are in the form of DNA and RNA and a...
How do you identify a phosphodiester bond?
A phosphodiester bond is identified as a bond between 2 sugar hydroxyl groups and a phosphate group. It is found in DNA and RNA.
How is a phosphodiester bond formed in DNA?
A phosphodiester bond is formed between two nucleotides to form the sugar-phosphate backbone of DNA. This reaction occurs as a condensation reactio...
How to understand phosphodiester bond?
To understand a phosphodiester bond, we first need to understand the basic structure of DNA and RNA. We know that DNA has a double helix structure, whereas RNA has a similar structure, except that it only has a single strand. DNA consists of three parts–a nitrogenous base, a sugar molecule and a triphosphate group.
What is the structure of DNA and RNA?
Structure of DNA and RNA. Nucleotide. Phosphodiester Bond. A phosphodiester bond is formed between two sugar molecules and a phosphate group. This bond connects nucleotides, which form the backbone of a DNA or RNA chain. DNA and RNA, as we know, are extremely important biomolecules found in living organisms. They are responsible for making us ...
What is the term for the time when a phosphoric acid molecule forms two ester bonds?
A phosphodiester bond literally refers to the time when a phosphoric acid molecule forms two ester bonds. As shown above, a nucleotide molecule already has one ester bond when the nucleoside attaches to the phosphate group. This phosphate group attaches with the sugar molecule of the neighboring nucleotide to link them.
What are the two parts of a nucleotide?
As mentioned above, a nucleotide molecule consists of 2 parts–a nucleoside and a phosphate group. Interlinked nucleotides form a single strand of genetic material. In the case of DNA, two strands are linked together by their nitrogenous bases to form the double-stranded structure.
How does phosphoric acid form a nucleotide?
Consider a phosphoric acid molecule. It has already lost one hydrogen atom to a sugar molecule to form a nucleotide. This phosphoric acid now undergoes a similar process to link with another sugar molecule. However, the difference is that when it links to the sugar molecule while forming the nucleotide, it attaches at the 5 th carbon of the ribose sugar. During bond formation between 2 different nucleotides, the hydroxyl group (-OH) is lost from the 3 rd carbon of the ribose sugar. The same process ensues—a hydrogen is lost from the phosphoric acid and an -OH is lost from the sugar to form a water molecule.
What is the name of the second ester bond?
This forms the second ester bond, so it gets the name phospho di ester bond. As mentioned, phosphodiester bonds can be in nucleotides containing a monophosphate, diphosphate or triphosphate.
What are the three parts of DNA?
DNA consists of three parts–a nitrogenous base, a sugar molecule and a triphosphate group. There are four nitrogenous bases: adenine, guanine, cytosine and thymine (uracil in RNA). The base, attached to the sugar molecule, is known as a nucleoside. The nucleoside attached to the phosphate group is called a nucleotide.
Should I study DNA replication?
I think you should first study DNA replication (if you haven't, yet) to get more thorough knowledge about the topic.
Is hydrogen in DNA?
Generally hydrogen is not shown in DNA structure. I have made a simple diagram to show where all hydrogens go:
Why is RNA hydrolyzed?
RNA is susceptible to this base-catalyzed hydrolysis because the ribose sugar in RNA has a hydroxyl group at the 2’ position. This feature makes RNA chemically unstable compared to DNA, ...
How does RNA hydrolysis occur?
RNA hydrolysis occurs when the deprotonated 2’ OH of the ribose, acting as a nucleophile, attacks the adjacent phosphorus in the phosphodiester bond of the sugar-phosphate backbone of the RNA. There is a transition state (shown above), where the phosphorus is bonded to five oxygen atoms. The phosphorus then detaches from the oxygen connecting it to the adjacent sugar, resulting in ester cleavage of the RNA backbone. (This mechanism is also referred to as RNA cleavage.) This produces a 2’,3’-cyclic phosphate that can then yield either a 2’- or a 3’-nucleotide when hydrolyzed. This process is shown in Figure 1.
What is the role of splicing ribozymes in RNA splicing?
Splicing ribozymes catalyze RNA splicing, removing a section of RNA that contains a mutation and replacing it with well-functioning RNA. Existing ribozymes can also be altered in a way that changes the reaction (s) that the ribozyme catalyzes.
What is the mechanism of base catalyzed RNA hydrolysis?
Mechanism of base catalyzed RNA hydrolysis. 1) Base-catalyzed deprotonation of the 2′-OH group, ena bling the deprotonated 2′ hydroxyl's nucleophilic attack on the adjacent phosphorus. 2) Transition state. 3) Phosphodiester bond is broken, cleaving the RNA backbone. 4) The 2′,3′-cyclic phosphate group (in step 3) hydrolyzes to either the 2′ or 3′ phosphate.
What is the process of cleavage of RNA?
This process is known as an auto-hydrolysis or a self-cleavage reaction. Spontaneous cleavage in an RNA molecule is much more likely to occur when it is single-stranded. Auto-hydrolysis or self-cleavage reactions take place in basic solutions, where free hydroxide ions in solution can easily deprotonate the 2’ OH of the ribose. This deprotonation makes the reaction base-catalyzed and increases spontaneity of the reaction.
What are the applications of ribozymes?
Applications include the use of ribozymes in gene therapy to control gene expression in bacteria and eukaryotes , and to inhibit viral replication. Hammerhead ribozymes, in particular, can be designed such that they will cleave a desired RNA. These ribozymes can be designed to prevent expression of a particular gene, for example.
What enzyme is used to cleave RNA?
Several different enzymes catalyze cleavage at specific sites on an RNA molecule. One such enzyme is Ribonuclease A (RNase A), a protein enzyme. RNase A contains histidine in its active site, and uses it to accomplish acid-base catalysis and cleavage of RNA.
What is the pKa of the phosphate group in DNA and RNA molecules?
But most of the phosphate groups in DNA and RNA molecules participate in phosphodiester bonds, thus only one of its hydroxyl groups are free to dissociate.
What is the OH group used for in nucleic acids?
In nucleic acids, two of the three OH-groups are used up for bonds to (desoxy)ribose, only one acidic group is left. Because of mesomery between the =O and -O - groups, the anion is stabilised and the p K a correspondingly low, some sources say 2, some even 0.
What equation is used to determine the phosphate group in DNA?
In order to understand the pKa of the phosphate groups in DNA, the Henderson-Hasselbalch equation and the intracellular pH must be taken into account.
What is the charge of phosphate groups in DNA?
The pKa of phosphate groups in DNA or RNA is 2 and gives a negative charge at neutral pH (pH=7). This charge-charge repulsion forces the phosphate groups to take opposite positions of the DNA strands and is neutralized by proteins (histones), metal ions such as magnesium and polyamines.
How many hydroxyl groups are in a phosphate?
(The exception are phosphates at the 5'-end, that have two hydroxyl groups.)
Where do phosphate groups come from in DNA?
Now, it is important to consider that the phosphate groups in DNA come from dNTPs. So you have to look at how this molecule comes about.
Does DNase remove DNA?
If I use DNase treatment (thermofisher AM1906) it only works on LiCl extracted RNA but does not efficiently remove DNA (from 5 to 1 ng/ul, after 2 hours doubling enzyme and buffer). With this conditions RNA starts to degrade.
What is the bond made by the water oxygen to the beta phosphoryl of ATP?
When you write it this way, it is clear that a bond is made by the water oxygen to the beta phosphoryl of ATP (bond making), this results in the release of the gamma phosphate (bond cleavage) [Yes, there is a deprotonation and reprotonation steps in the actual mechanism]. However, this is only half the story. The phosphoryls on ATP are each negatively charged at physiological pH, so one expects charge-charge repulsion within the ATP molecule. When hydrolyzed, the phosphate and the ADP are free to go their separate ways and that relieves the charge-charge repulsion. Further, both the ADP and the phosphate are able to (nearly) independently occupy the volume of solution in which they are found and that adds an entropic factor to the free energy of hydrolysis. Complicating things a bit more is the fact that you rarely (never in the cell) have just ATP, but you have M g 2 + A T P and M g 2 + A D P but the above story still holds. I mention this just for completeness, and so that you will realize that computing these sorts of energies precisely is difficult. This is why we use experimentally derived tables [ 1].
Why is more ATP produced from NADH than from FADH2?
The reason why more ATP are produced from NADH than from FADH2 is that FAD takes less energy to reduce than does NAD+; so when the opposite (oxidation) occur s, more energy is released from NADH than from FADH2.
How much energy is produced when NADH is oxidized?
When NADH is oxidized, sufficient energy is contributed to the proton gradient for the phosphorylation of nearly 3 ATP (about 2.5 to be more precise) by ATP synthase. Similarly, when FADH2 is oxidized, nearly 2 ATP are formed. This difference might be due to the difference in the complexes in which they are putting their electrons. Complex 1 is in higher energy state than complex 2, so it provides more energy.
How does FADH2 work?
Here, we can see that FADH2 provides electrons to complex 2 which in turn passes it to another complex, eventually forming water and CO2. These complexes utilize the energy of passing electrons to send the protons ( H+) from mitochondrial matrix to inter-membrane space creating proton gradient. When the protons are allowed to come back to the matrix following its gradient via ATP synthase (blue pump in the figure), ATP synthase uses the energy of proton passing through it to form ATP.
How does ATP synthase work?
A portion of this section of ATP synthase spins (rotates) with the movement of protons through the membrane. The dynamic electrostatic attractions that appear during this rotation cause the ATP Synthase catalytic nucleotide binding sites to perform a series of physical conformational changes that leads to ATP synthesis. Energy is generated as positive hydrogen ions (protons) that move down a formed electrochemical gradient from the outer membranes of the mitochondria to its inner structure, eerily known as The Matrix (or similarly from outer membrane to inside in chloroplasts for plants). This gradient must be positive to negative to move the proton down it. Since it is a gradient, it must have an initial repulsive effect on protons at the positive end, and an attractive effect at the other end, where the electrons have been transported.
What is the model of ATP synthesis?
Naturally, like most other scientific disciplines, there is an accepted model for the process of ATP synthesis. It is called the alternating catalytic model (which you are obviously familiar with). According to this model, the force across the inner mitochondrial membrane used to move protons is generated by the electron transport chain and is the engine for the movement of protons through the membrane via an active area (
Why does ATP not go forward?
But the reason that the reaction does not go forward spontaneously is because there is a kinetic barrier preventing the hydrolysis. I like to compare it to a ball sitting on top of a cone with the top rounded out a bit like this: