Procedure of Restriction Digestion of DNA
- Always keep restriction enzyme (EcoRI or HindIII), substrate (lambda DNA), and assay buffer in an ice bucket.
- Take 2-5 μg of the lambda DNA as substrate in an Eppendorf tube and dissolve it in an appropriate volume of water.
- Add ul of about 10X assay buffer (available with the restriction enzyme) to the DNA in the Eppendorf tube,...
What are the types of restriction enzymes?
Type I Restriction Enzymes
- Type I restriction enzymes possess both restriction and modification activities. ...
- Cleavage takes place nearly 1000 base pairs away from the restriction site.
- The structure of the recognition site is asymmetrical. ...
- For their function, the type I restriction enzymes require S- adenosylmethionine (SAM), ATP, and Mg 2+
What are restriction enzymes purpose?
Restriction enzymes are found in bacteria which are actually meant for Host controlled restriction which is helpful in protecting the bacterial DNA by phage invasion. Such restriction enzymes digest the phage DNA by cleaving it at internal positions. Bacterial DNA are methylated so they are immune to the action of these restriction enzymes.
How many restriction enzymes are there?
There are three types of Restriction Enzymes: Type I, Type II, and Type III. Type I restriction enzymes are also called restriction endonucleases. They are made of two long strands of DNA joined together. These restriction enzymes recognize certain sequences of DNA and cleave them at a site.
Do humans have restriction enzymes?
The HsaI restriction enzyme from the embryos of human, Homo sapiens, has been isolated with both the tissue extract and nuclear extract. It proves to be an unusual enzyme, clearly related functionally to Type II endonuclease. HsaI seems to be an isoschizomer of EcoRI, but it has a distinctive property of elution, differing from EcoRI.

How do restriction enzymes digest?
Restriction digestion is accomplished by incubation of the target DNA molecule with restriction enzymes - enzymes that recognize and bind specific DNA sequences and cleave at specific nucleotides either within the recognition sequence or outside of the recognition sequence.
How do restriction enzymes break DNA?
Like all enzymes, a restriction enzyme works by shape-to-shape matching. When it comes into contact with a DNA sequence with a shape that matches a part of the enzyme, called the recognition site, it wraps around the DNA and causes a break in both strands of the DNA molecule.
What does the restriction enzymes actually do to the DNA?
Restriction enzymes, also called restriction endonucleases, recognize a specific sequence of nucleotides in double stranded DNA and cut the DNA at a specific location. They are indispensable to the isolation of genes and the construction of cloned DNA molecules.
What is DNA restriction digest?
Restriction Digestion is the process of cutting DNA molecules into smaller pieces with special enzymes called Restriction Endonucleases (sometimes just called Restriction Enzymes or RE's).
How do restriction enzymes work quizlet?
how does a Restriction enzyme work: it cuts double stranded DNA somewhere in the middle; either at or near the recognition site and are then isolated from bacterial sources. - they carry both modification, i.e., methylation, and restriction, i.e., cleavage activities in the same protein.
What bonds do restriction enzymes break?
Restriction enzymes break the phosphodiester bond between the phosphate and the pentose sugar in the sugar-phosphate backbone at the specific site.
What are restriction enzymes how do they work what are recognition sites?
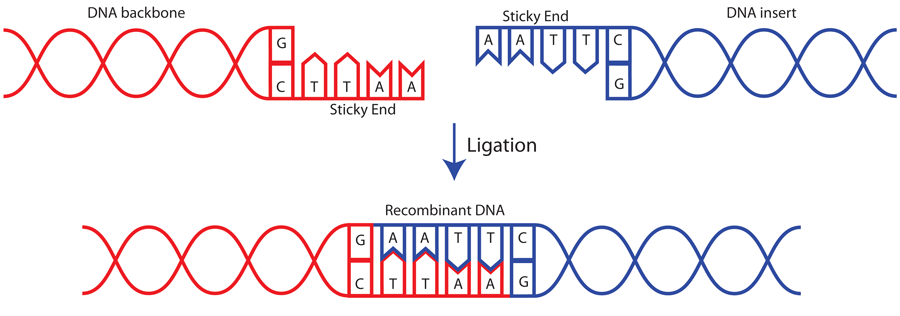
A restriction enzyme is a DNA-cutting enzyme that recognizes specific sites in DNA. Many restriction enzymes make staggered cuts at or near their recognition sites, producing ends with a single-stranded overhang. If two DNA molecules have matching ends, they can be joined by the enzyme DNA ligase.
How do restriction enzymes work how can they help to determine variation in DNA sequences?
When a restriction enzyme cleaves a restriction site, the reaction creates highly reactive "sticky ends" on the broken DNA. This is useful to the biotechnologist! By cutting open vector DNA with the same with restriction enzymes used to cleave the target DNA, complementary "sticky ends" are created.
What are two functions of restriction enzymes?
The function of restriction endonucleases is mainly protection against foreign genetic material especially against bacteriophage DNA. The other functions attributed to these enzymes are recombination and transposition.
What enzyme digests DNA?
restriction endonucleaserestriction enzyme, also called restriction endonuclease, a protein produced by bacteria that cleaves DNA at specific sites along the molecule. In the bacterial cell, restriction enzymes cleave foreign DNA, thus eliminating infecting organisms.
Can DNA be completely digested by a restriction enzyme?
One unit of restriction endonuclease completely digests 1 µg of substrate DNA in 1 hour. However, supercoiled plasmid DNA generally requires more than 1 unit/µg to be digested completely. Most researchers add a 10-fold excess of enzyme to their reactions in order to ensure complete cleavage.
How do restriction enzymes work how can they help to determine variation in DNA sequences?
When a restriction enzyme cleaves a restriction site, the reaction creates highly reactive "sticky ends" on the broken DNA. This is useful to the biotechnologist! By cutting open vector DNA with the same with restriction enzymes used to cleave the target DNA, complementary "sticky ends" are created.
What cuts DNA into small fragments?
Restriction enzymes are DNA-cutting enzymes. Each enzyme recognizes one or a few target sequences and cuts DNA at or near those sequences. Many restriction enzymes make staggered cuts, producing ends with single-stranded DNA overhangs.
Why do restriction enzymes cut DNA into different sized fragments?
A DNA segment, digested by a specific restriction enzyme, is cut into smaller DNA fragments of different sizes depending on the number and location of the recognition sites present within the DNA sequence.
How the DNA is cut?
To cut DNA at known locations, researchers use restriction enzymes that have been purified from various bacterial species, and which can be purchased from various commercial sources. These enzymes are usually named after the bacterium from which they were first isolated.
What is restriction digestion?
The restriction endonucleases are involved in the bacterial and prokaryotic defence mechanism and hence the restriction digestion too. The virus and phages always target bacterial DNA.
What is DNA digestion?
The process of cutting or cleaving DNA into smaller fragments with the help of the chemicals, physical and enzymatic methods are called the process of the DNA digestion.
What digestion system helps the bacterial own DNA from the phage invasion and from the other virus?
The restriction endonuclease digestion system helps the bacterial own DNA from the phage invasion and from the other virus.
What enzyme is used to map polymorphism?
The process of cleaving DNA at a particular location with the help of the specific type of restriction endonuclease enzyme which helps in mapping, polymorphism study and studying mutation called as restriction digestion.
What are the sequences of DNA in a sense strand from 5’ to 3’ and antisense?
The sequences of DNA in a sense strand from 5’ to 3’ and antisense strand from 3’ to 5’, are same, these types of DNA sequences are the palindromic sequences. See the figure, The restriction endonuclease digestion system helps the bacterial own DNA from the phage invasion and from the other virus.
Why is water important in restriction digestion?
Nuclease-free water: water is one of the important ingredients in restriction digestion procedure for balancing the ionic strength and the reaction. But, the water used into the restriction digestion must be nuclease-free, otherwise, other nucleases cut DNA randomly leads to false positive results.
What is the best method for DNA extraction?
The best method for DNA extraction for the restriction digestion is phenol-chloroform D NA extraction method, an enzymatic method of DNA extraction or you can use the ready to use DNA extraction kits. Choose one of the methods of DNA extraction and extract DNA with ~1.80 purity having a good quantity of DNA.
Why are restriction enzymes important?
Restriction enzymes have been used to help produce vaccines, pharmaceutical products, insect resistant crops, and a host of other products.
What is restriction enzyme?
Restriction enzymes are enzymes that cut DNA into fragments based upon recognizing a specific sequence of nucleotides. Restriction enzymes are also known as restriction endonucleases. Regina Bailey is a board-certified registered nurse, science writer and educator. Her work has been featured in "Kaplan AP Biology" and "The Internet for Cellular ...
What Is DNA Ligase?
The sticky ends of the fragments produced by restriction enzymes are useful in a laboratory setting. They can be used to join DNA fragments from both different sources and different organisms. The fragments are held together by hydrogen bonds. From a chemical perspective, hydrogen bonds are weak attractions and are not permanent. Using another type of enzyme however, the bonds can be made permanent.
What sequences do restriction enzymes recognize?
For example, a restriction enzyme may recognize a specific sequence of guanine, adenine, adenine, thymine, thymine, cytosine. When this sequence is present, the enzyme can make staggered cuts in the sugar-phosphate backbone in the sequence. But if restriction enzymes cut based on a certain sequence, how do cells like bacteria protect their own DNA ...
What happens when restriction enzyme cuts DNA?
Since the DNA is cut on both strands, there will be complementary ends that can hydrogen bond to one another. These ends are often called "sticky ends."
What enzymes dismantle DNA?
Restriction enzymes dismantle foreign DNA by cutting it into fragments. This disassembling process is called restriction.
Why are methyl groups added to DNA?
In a typical cell, methyl groups (CH 3) are added to the bases in the sequence to prevent recognition by the restriction enzymes. This process is carried out by complementary enzymes that recognize the same sequence of nucleotide bases as restriction enzymes. The methylation of DNA is known as modification. With the processes of modification and ...
What enzyme is used to digest bacteriophage lambda DNA?
Digestion of bacteriophage lambda (λ) DNA using a restriction enzyme.
What enzymes should be kept in an ice bucket?
Always keep restriction enzyme (EcoRI or HindIII), substrate (lambda DNA ), and assay buffer in an ice bucket.
How many DNA fragments are there in lambda?
The exact number and size of the bands obtained depend on the restriction enzymes used for digesting the lambda DNA (sample ‘B’). As explained earlier, 6 DNA fragments ( 21, 226, 7421, 5804, 4878, and 3530 bp) are observed if EcoRI is used and 8 DNA fragments (23, 130, 9416, 6557, 4361, 2322, 2027, 564, and 125 bp) are observed after using HindIII.
How long to stop reaction in Eppendorf tube?
After one hour stop the reaction by addition of 3.33 μl of 6X gel loading buffer to the Eppendorf tube. Label the vial as ‘A’ and put it on ice.
Where are restriction enzymes found?
Restriction enzymes are found in bacteria (and other prokaryotes). They recognize and bind to specific sequences of DNA, called restriction sites. Each restriction enzyme recognizes just one or a few restriction sites. When it finds its target sequence, a restriction enzyme will make a double-stranded cut in the DNA molecule.
What is a restriction enzyme?
A restriction enzyme is a DNA-cutting enzyme that recognizes specific sites in DNA. Many restriction enzymes make staggered cuts at or near their recognition sites, producing ends with a single-stranded overhang. If two DNA molecules have matching ends, they can be joined by the enzyme DNA ligase.
What is the enzyme that seals the gap between two DNA molecules?
If two DNA molecules have matching ends, they can be joined by the enzyme DNA ligase. DNA ligase seals the gap between the molecules, forming a single piece of DNA. Restriction enzymes and DNA ligase are often used to insert genes and other pieces of DNA into plasmids during DNA cloning.
How is recombinant plasmid produced?
Right: recombinant plasmid produced when gene goes in backwards ("pointing" back towards the promoter that is already in the plasmid). Restriction digests and ligations like this one are performed using many copies of plasmid and gene DNA. In fact, billions of molecules of DNA are used in a single ligation!
Why are blunt-ended fragments harder to ligate together?
However, blunt-ended fragments are harder to ligate together (the ligation reaction is less efficient and more likely to fail) because there are no single-stranded overhangs to hold the DNA molecules in position.
How does DNA ligase work?
How does DNA ligase do this? Using ATP as an energy source, ligase catalyzes a reaction in which the phosphate group sticking off the 5’ end of one DNA strand is linked to the hydroxyl group sticking off the 3’ end of the other. This reaction produces an intact sugar-phosphate backbone.
Why do enzymes leave sticky ends?
Sticky ends are helpful in cloning because they hold two pieces of DNA together so they can be linked by DNA ligase. Not all restriction enzymes produce sticky ends.
What is restriction enzyme?
Restriction enzymes are the backbone reagents of cloning, but are used in clinical applications associated with fingerprinting – genetic identity, epidemiology, and in preparation for blotting for other applications. Cloning Step 1: Restriction digestion. This is a step that essentially cuts DNA into little bits.
What is the purpose of restriction digestion?
Once the DNA has undergone restriction digestion, it may be used to recombine with any other piece of DNA that has the complementary ends, regardless of the source of that DNA. This allows insertion of a segment of DNA into a plasmid that has been cleaved with the same enzyme (s).
How does electrophoresis work?
Electrophoresis is a method whereby charged molecules in solution migrate in response to an electric field. Because of the phosphate backbone in DNA molecules, they have a net negative charge and migrate toward the anode (positive pole) in an electric field. Their rate of migration or mobility is related to the strength of the field, the size of the molecule, as well as the medium (gel) in which they are migrating. For separation of DNA molecules, electrophoresis is often carried out in a horizontal apparatus containing a gel made of agarose. Agarose is a highly purified polysaccharide derived from agar (seaweed) that is not contaminated with charged material. It comes in powder form and is dissolved by boiling in aqueous solutions. It remains in a liquid state until the temperature is lowered to about 40ºC at which point the agar solution gels. The gel is stable and will not dissolve again until raised back to boiling temperatures. The pore size of the agarose is adjusted by changing the concentration of the agarose in the gel. The higher the concentration of agarose, the smaller the gel pore size. Working concentrations are usually in the range of 0.5% to 2.0%. A 2.0% gel has a smaller pore size than a 0.5% gel and will allow better separation of short DNA molecules (those that are a few hundred base pairs in length) than a 1.0% gel which might be used to separate molecules that are larger in size (thousands of base pairs in length). The migration of molecules in agarose is size-dependent and allows separation of molecules up to about 20 kilobases (kb) in size. The smaller the molecule, the more rapidly it will be able to pass through the pores in the agarose in its migration toward the positive pole. Thus, in a mixture of DNA molecules of different sizes, the shortest fragments will migrate the most rapidly, while the largest will be retarded to the greatest extent by the pores in the agarose and will migrate the most slowly. The best separation is achieved after experimenting with variations in the concentration of agarose and the separation time in the electric field. When the agarose gel is prepared, a “slot maker” or comb is inserted into the chamber so that small wells or slots are formed when the agarose gels or sets. The DNA sample to be analyzed is first mixed with a glycerol-dye solution. The glycerol in the sample makes it more dense than the running buffer so that it can be applied into the sample slot of a submerged gel in an electrophoretic chamber without diffusing away prior to application of electrical current. The inclusion of the dye (bromophenol blue or others) helps to make the sample visible during application and provides a marker to help track the progress of the electrophoretic run because it is also negatively charged and migrates toward the positive pole at a rate similar to that of small DNA molecules. To better visualize the separation of DNA molecules during and after the electrophoretic run, ethidium bromide, SYBR green or SYBR gold are included in low concentrations (0.5 μg/ml) in the gel and in the running buffer, or in the loading dye itself (SYBRs). SYBRs bind to double-stranded DNA and becomes fluorescent when excited by ultraviolet (UV) light. A DNA ladder that contains DNA molecules of defined length is also included as one of the samples in one of the lanes of the gel to provide an internal marker of DNA fragment sizes and to provide an indication of how well molecules have separated in the gel. The DNA bands can be visualized and photographed by placing the agarose gel on a UV light box. Care must be taken to wear a UV resistant shield, goggles or glasses when viewing gels on a UV box to avoid damage to your eyes. In addition, ethidium bromide is a carcinogen and mutagen and care must be taken to avoid skin contact with ethidium bromide- containing solutions. Always wear latex gloves when working with DNA samples during electrophoretic procedures to prevent contact with ethidium bromide in the gel or running buffer. Other dyes include SYBR green or gold, which are much safer to use and have similar binding characteristics as ethidium bromide, but in some circumstances, are not quite as sensitive in gels. To confirm the identity of a PCR or RT-PCR product, the agarose gel from above can be subjected to the same non-amplification techniques (blotting) as will be presented in Module 4.
What is the process of agarose gel electrophoresis?
Agarose Gel Electrophoresis. Electrophoresis is a method whereby charged molecules in solution migrate in response to an electric field. Because of the phosphate backbone in DNA molecules, they have a net negative charge and migrate toward the anode (positive pole) in an electric field.
How are nucleic acids separated?
Usually prior to blotting the DNA, RNA, or protein molecules for further studies and detection, nucleic acids are separated based on size or mass by electrophoresis. Like total protein electrophoresis, DNA and RNA can be separated by size/mass, but charge is irrelevant (DNA and RNA are negatively charged).
Why is cloning a gene sequence important?
Cloning of a gene sequence (or part) is also required to have stable templates for the production of probes, or to save that interesting PCR product. Long term storage of PCR products without cloning them, leaves them susceptible to degradation by bacterial exonucleases (enzymes that digest DNA from the ends).
What are the sites of restriction?
The sites on the DNA molecule recognized by the enzymes are called restriction sites. Restrictions sites are palindromic, reading the same 5’?3’ on either strand of the DNA. The various enzymes are named for the bacteria from which they have been derived. Eco is derived from Escherichia coli and Hin from Haemophilus influenzae, for example. You will note that the first panel above the cut leaves no overhang and is called a blunt cut leaving blunt ends. Any blunt end can be rejoined to any other blunt end – something to remember when cloning PCR products. The other panel shows a cut that leaves overhanging ends called either 5’ or 3’ overhangs depending on which strand is overhanging. These overhanging ends are also called “sticky ends”.
How long to digest plasmid?
Mix by flicking, then tap on bench to get liquid to bottom of tube. Incubate at correct temperature for approx. 2 hours. (1 - 4 hr is OK, but overnight digest is too long; this can lead to ‘raggedy ends’ of the plasmid even if it looks OK on a gel; this is due to non-specific nuclease activity)
How to thaw plasmids?
Retrieve the plasmid from the freezer, allow to thaw, (e.g. in 37°C water bath, or rub in your hands, or on bench etc), then put it on ice when it is thawed. Its not good to leave the plasmid stock at room temp or above for prolonged periods or it may degrade due to traces of nucleases.
How many ng of DNA is in a sample?
Range may be 100-1000 ng of each DNA, in a volume of 30 – 200 µl.
Can you leave plasmids in the freezer?
in 37°C water bath, or rub in your hands, or on bench etc), then put it on ice when it is thawed. Its not good to leave the plasmid stock at room temp or above for prolonged periods or it may degrade due to traces of nucleases.
Do you need to buy reagents for restriction enzyme digest?
This step prevents the vector religating to itself; it is not required (in theory!) when you are cloning with two different restriction enzymes. Only buy the reagents to perform this step if you Really need to do a single restriction enzyme digest.
Do you thaw glycerol?
These are often extremely heat sensitive. They will be in glycerol stock and thus do not need hand thawing. Move immediately from the freezer to the ice box.
Can you proceed to the Ligation Protocol?
You can now proceed to the Ligation Protocol, but the chance of success is quite low. It can be significantly improved by adding either an inactivation or a purification step here.
How many units of DNA digest?
10 units, i.e 1uL is usually enough to digest 1ug of DNA.
Can you use DNA as a cloning vector?
If you are going to use you DNA as a cloning vector, some over-di gestion might be required to ensure absence of partially digested DNA, which may produce empty clones after ligation. Cite. 1 Recommendation. 21st Oct, 2016.
