
Restriction Enzymes: Nature's Scissors
- Restriction enzymes are naturally produced in bacteria as a defense mechanism againt foreign DNA.
- Often called ‘molecular scissors’ since they can ‘cut’ DNA.
- Each restriction enzyme cuts a specific nucleotide sequence of DNA.
- These enzymes are now used a genetic tool for manipulating DNA and they have implications many areas of biotechnology.
What is the function of restriction enzymes?
What is the natural function of a restriction enzyme? Their natural function is to destroy foreign DNA entering the cell by cleaving the bacteriophage DNA to prevent infection. The cell's own DNA is modified by methylation to protect it from its own enzyme. Each restriction enzyme has a specific methylase.
Do restriction enzymes cut DNA?
Restriction enzymes are a class of enzymes that cut DNA into fragments based upon recognizing a specific sequence of nucleotides. Restriction enzymes are also known as restriction endonucleases. While there are hundreds of different restriction enzymes, they all work in essentially the same way.
How many restriction enzymes are there?
There are three types of Restriction Enzymes: Type I, Type II, and Type III. Type I restriction enzymes are also called restriction endonucleases. They are made of two long strands of DNA joined together. These restriction enzymes recognize certain sequences of DNA and cleave them at a site.
How do restriction endonucleases work?
How Do Restriction Endonucleases Work Brainly? Restriction enzyme, also called restriction endonuclease, is a protein produced by bacteria that cleaves DNA at specific sites along the molecule. Restriction endonucleases cut the DNA double helix in very precise ways. It cleaves DNA into fragments at or near specific recognition sites within the ...
How do restriction enzymes function?
A restriction enzyme is a protein isolated from bacteria that cleaves DNA sequences at sequence-specific sites, producing DNA fragments with a known sequence at each end. The use of restriction enzymes is critical to certain laboratory methods, including recombinant DNA technology and genetic engineering.
How do restriction enzymes cut DNA?
Restriction enzymes cut DNA bonds between 3′ OH of one nucleotide and 5′ phosphate of the next one at the specific restriction site. Adding methyl groups to certain bases at the recognition sites on the bacterial DNA blocks the restriction enzyme to bind and protects the bacterial DNA from being cut by themselves.
How do restriction enzymes work quizlet?
how does a Restriction enzyme work: it cuts double stranded DNA somewhere in the middle; either at or near the recognition site and are then isolated from bacterial sources. - they carry both modification, i.e., methylation, and restriction, i.e., cleavage activities in the same protein.
How do restriction enzymes work in recombinant DNA?
A restriction enzyme is a DNA-cutting enzyme that recognizes specific sites in DNA. Many restriction enzymes make staggered cuts at or near their recognition sites, producing ends with a single-stranded overhang. If two DNA molecules have matching ends, they can be joined by the enzyme DNA ligase.
Where do restriction enzymes cleave DNA?
Restriction enzymes cleave DNA at specific nucleotide sequences. Restriction endonucleases cleave double-stranded DNA. These enzymes are sequence specific, and each enzyme acts at a limited number of sites in DNA called recognition, or cutting, sites.
What do restriction enzymes restrict?
The restriction enzyme prevents replication of the phage DNA by cutting it into many pieces. Restriction enzymes were named for their ability to restrict, or limit, the number of strains of bacteriophage that can infect bacteria.
How do restriction enzymes cut DNA quizlet?
Terms in this set (44) How do restriction enzymes cut DNA sequences? They cut DNA at sites, called recognition sites, that have specific nucleotide sequences.
What do restriction enzymes do to DNA quizlet?
What is the function of a restriction enzyme? they recognize specific sequences in DNA and then cut the DNA and then cut the DNA to produce fragments, called restriction fragments.
What are restriction enzymes how do they work what are recognition sites?
How do restriction enzymes work? Like all enzymes, a restriction enzyme works by shape-to-shape matching. When it comes into contact with a DNA sequence with a shape that matches a part of the enzyme, called the recognition site, it wraps around the DNA and causes a break in both strands of the DNA molecule.
Why do restriction enzymes not cut bacterial DNA?
Bacterial DNA is highly methylated and is unrecognizable for the restriction enzymes, thus being prevented from cleavage.
How the DNA is cut?
To cut DNA at known locations, researchers use restriction enzymes that have been purified from various bacterial species, and which can be purchased from various commercial sources. These enzymes are usually named after the bacterium from which they were first isolated.
What is the process of cutting DNA into fragments?
Terms in this set (5)genetic engineering. the process of cutting DNA into fragments and inserting the fragments with a desired gene into another organism of the same or different species.recombinant DNA. ... restriction enzymes. ... transgenic organism. ... vector.
Do restriction enzymes cut DNA randomly?
Type I restriction enzymes cut DNA at random locations far from their recognition sequence, type II cut within or close to their recognition sequence, type III cut outside of their recognition sequence, and type IV typically recognize a modified recognition sequence.
What enzyme cuts DNA into fragments?
A restriction enzyme, restriction endonuclease, or restrictase is an enzyme that cleaves DNA into fragments at or near specific recognition sites within molecules known as restriction sites.
Where Do Restriction Enzymes Come from?
Restriction enzymes999 are found in bacteria. Bacteria55101010 use restriction enzymes111111 to kill viruses – the enzymes attack the viral DNA and...
How Do Restriction Enzymes Work?
Like all enzymes, a restriction enzyme works by shape-to-shape matching. When it comes into contact with a DNA sequence with a shape that matches a...
When Are Restriction Enzymes used?
Restriction enzymes are a basic tool for biotechnology research. They are used for DNA cloning and DNA fingerprinting.
Different Types of Restriction Enzyme
Scientists have identified and purified hundreds of different types of restriction enzymes. They are named after the genus and species of the organ...
DNA Fragments: Blunt Or Sticky Ends?
DNA consists of two complementary strands of nucleotides202020 that spiral around each other in a double helix. Restriction enzymes cut through bot...
How does a bacterium use a restriction enzyme?
A bacterium uses a restriction enzyme to defend against bacterial viruses called bacteriophages, or phages. When a phage infects a bacterium, it inserts its DNA into the bacterial cell so that it might be replicated. The restriction enzyme prevents replication of the phage DNA by cutting it into many pieces. Restriction enzymes were named ...
Why are restriction enzymes used in genetic engineering?
Restriction enzymes can be isolated from bacterial cells and used in the laboratory to manipulate fragments of DNA, such as those that contain genes; for this reason they are indispensible tools of recombinant DNA technology ( genetic engineering ).
How does type II restriction enzyme differ from type I?
Type II restriction enzymes also differ from types I and III in that they cleave DNA at specific sites within the recognition site; the others cleave DNA randomly, sometimes hundreds of bases from the recognition sequence.
What enzymes prevent replication of phage DNA?
The restriction enzyme prevents replication of the phage DNA by cutting it into many pieces. Restriction enzymes were named for their ability to restrict, or limit, the number of strains of bacteriophage that can infect a bacterium.
How many bases are there in a type II restriction enzyme?
These enzymes recognize a few hundred distinct sequences, generally four to eight bases in length. Type IV restriction enzymes cleave only methylated DNA and show weak sequence specificity.
Which type of enzyme is independent of its methylase?
Types I and III enzymes are similar in that both restriction and methylase activities are carried out by one large enzyme complex, in contrast to the type II system, in which the restriction enzyme is independent of its methylase.
When were restriction enzymes discovered?
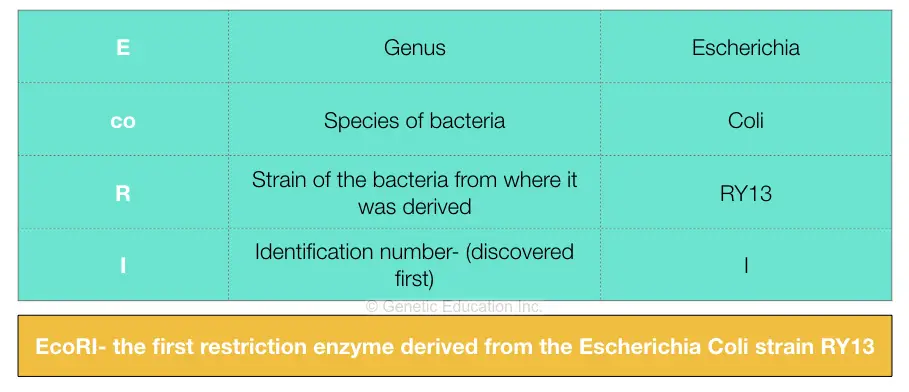
Restriction enzymes were discovered and characterized in the late 1960s and early 1970s by molecular biologists Werner Arber, Hamilton O. Smith, and Daniel Nathans. The ability of the enzymes to cut DNA at precise locations enabled researchers to isolate gene-containing fragments and recombine them with other molecules of DNA—i.e., to clone genes. The names of restriction enzymes are derived from the genus, species, and strain designations of the bacteria that produce them; for example, the enzyme Eco RI is produced by Escherichia coli strain RY13. It is thought that restriction enzymes originated from a common ancestral protein and evolved to recognize specific sequences through processes such as genetic recombination and gene amplification.
How are artificial restriction enzymes generated?
Artificial restriction enzymes can be generated by fusing a natural or engineered DNA-binding domain to a nuclease domain (often the cleavage domain of the type IIS restriction enzyme Fok I ). Such artificial restriction enzymes can target large DNA sites (up to 36 bp) and can be engineered to bind to desired DNA sequences. Zinc finger nucleases are the most commonly used artificial restriction enzymes and are generally used in genetic engineering applications, but can also be used for more standard gene cloning applications. Other artificial restriction enzymes are based on the DNA binding domain of TAL effectors.
How do restriction enzymes recognize nucleotides?
Restriction enzymes recognize a specific sequence of nucleotides and produce a double-stranded cut in the DNA. The recognition sequences can also be classified by the number of bases in its recognition site, usually between 4 and 8 bases, and the number of bases in the sequence will determine how often the site will appear by chance in any given genome, e.g., a 4-base pair sequence would theoretically occur once every 4^4 or 256bp, 6 bases, 4^6 or 4,096bp, and 8 bases would be 4^8 or 65,536bp. Many of them are palindromic, meaning the base sequence reads the same backwards and forwards. In theory, there are two types of palindromic sequences that can be possible in DNA. The mirror-like palindrome is similar to those found in ordinary text, in which a sequence reads the same forward and backward on a single strand of DNA, as in GTAATG. The inverted repeat palindrome is also a sequence that reads the same forward and backward, but the forward and backward sequences are found in complementary DNA strands (i.e., of double-stranded DNA), as in GTATAC (GTATAC being complementary to CATATG). Inverted repeat palindromes are more common and have greater biological importance than mirror-like palindromes.
How do type II enzymes differ from type I enzymes?
They form homodimers, with recognition sites that are usually undivided and palindromic and 4–8 nucleotides in length. They recognize and cleave DNA at the same site, and they do not use ATP or AdoMet for their activity— they usually require only Mg 2+ as a cofactor. These enzymes cleave the phosphodiester bond of double helix DNA. It can either cleave at the center of both strands to yield a blunt end, or at a staggered position leaving overhangs called sticky ends. These are the most commonly available and used restriction enzymes. In the 1990s and early 2000s, new enzymes from this family were discovered that did not follow all the classical criteria of this enzyme class, and new subfamily nomenclature was developed to divide this large family into subcategories based on deviations from typical characteristics of type II enzymes. These subgroups are defined using a letter suffix.
What are the different types of restriction endonucleases?
Types. Naturally occurring restriction endonucleases are categorized into four groups (Type s I, II III, and IV) based on their composition and enzyme cofactor requirements, the nature of their target sequence, and the position of their DNA cleavage site relative to the target sequence.
How did restriction endonuclease evolve?
In addition, there is mounting evidence that restriction endonucleases evolved as a selfish genetic element.
What are different restriction enzymes that recognize the same sequence called?
Different restriction enzymes that recognize the same sequence are known as neoschizomers . These often cleave in different locales of the sequence. Different enzymes that recognize and cleave in the same location are known as isoschizomers .
What is the enzyme that cuts DNA?
A restriction enzyme, restriction endonuclease, or restrictase is an enzyme that cleaves DNA into fragments at or near specific recognition sites within molecules known as restriction sites. Restriction enzymes are one class of the broader endonuclease group of enzymes. Restriction enzymes are commonly classified into five types, which differ in their structure and whether they cut their DNA substrate at their recognition site, or if the recognition and cleavage sites are separate from one another. To cut DNA, all restriction enzymes make two incisions, once through each sugar-phosphate backbone (i.e. each strand) of the DNA double helix .
What are Restriction Enzymes?
The restriction enzyme is a protein produced by bacteria that cleaves the DNA at specific sites. This site is known as the restriction site.
Why are restriction enzymes important?
Restriction enzymes are important tools for genetic engineering. They can be isolated from the bacteria and used in the laboratories.
What enzyme cuts plasmids?
Restriction enzymes cut the plasmid producing single-stranded overhangs. The two DNA molecules are ligated with the help of DNA ligase to form a single DNA molecule. For more information on Restriction Enzymes, its types, applications and related topics, visit us @ BYJU’S Biology.
What are the two types of cuts that restriction enzymes produce?
The restriction enzymes generate two different types of cuts. Blunt ends are produced when they cut the DNA at the centre of the recognition sequence, and sticky ends produce an overhang.
What type of enzyme cuts DNA?
Type I. These restriction enzymes cut the DNA far from the recognition sequences. However, they do not produce discrete restriction fragments, hence, are of not much practical value. These are complex, multi-subunit restriction and modification enzymes.
Why are RFLP fragments used?
They are used in RFLP techniques to cut the DNA into smaller fragments to study the fragment length differences among the individuals.
How does bacteria prevent DNA degradation?
The bacteria prevents its own DNA sequences from degradation by the addition of the methyl group at the adenine or cytosine bases within the recognition sequence with the help of enzyme methylases. Also read: Cloning Vector.
What is restriction enzyme?
Restriction enzymes are enzymes that cut DNA into fragments based upon recognizing a specific sequence of nucleotides. Restriction enzymes are also known as restriction endonucleases. Regina Bailey is a board-certified registered nurse, science writer and educator. Her work has been featured in "Kaplan AP Biology" and "The Internet for Cellular ...
Why are restriction enzymes important?
Restriction enzymes have been used to help produce vaccines, pharmaceutical products, insect resistant crops, and a host of other products.
What Is DNA Ligase?
The sticky ends of the fragments produced by restriction enzymes are useful in a laboratory setting. They can be used to join DNA fragments from both different sources and different organisms. The fragments are held together by hydrogen bonds. From a chemical perspective, hydrogen bonds are weak attractions and are not permanent. Using another type of enzyme however, the bonds can be made permanent.
What sequences do restriction enzymes recognize?
For example, a restriction enzyme may recognize a specific sequence of guanine, adenine, adenine, thymine, thymine, cytosine. When this sequence is present, the enzyme can make staggered cuts in the sugar-phosphate backbone in the sequence. But if restriction enzymes cut based on a certain sequence, how do cells like bacteria protect their own DNA ...
What happens when restriction enzyme cuts DNA?
Since the DNA is cut on both strands, there will be complementary ends that can hydrogen bond to one another. These ends are often called "sticky ends."
What enzymes dismantle DNA?
Restriction enzymes dismantle foreign DNA by cutting it into fragments. This disassembling process is called restriction.
Why are methyl groups added to DNA?
In a typical cell, methyl groups (CH 3) are added to the bases in the sequence to prevent recognition by the restriction enzymes. This process is carried out by complementary enzymes that recognize the same sequence of nucleotide bases as restriction enzymes. The methylation of DNA is known as modification. With the processes of modification and ...
How Do Restriction Enzymes Work?
All restriction enzymes recognize a sequence of four- to eight-nucleotides called the recognition site. The nucleotide sequence is palindromic, meaning the sequence of ATCG’s read the same forwards on one strand and ‘backwards’ on the complementary strand. Look at the image to the right, do you recognize the palindrome? You’ll notice they read the same in the 5′ to 3′ direction on one strand as they do in the 5′ to 3′ direction on the complementary strand (a).
What is the function of restriction enzymes?
Restriction endonucleases – or restriction enzymes – recognize specific DNA sequences and cut them in a predictable manner. They are naturally produced by bacteria as a defense mechanism against foreign DNA. For use in botechnology, restriction enzymes are isolated from bacteria to be used a genetic tool in biotechnology.
Why do bacteria produce restriction enzymes?
Bacteria naturally produce restriction enzymes (also called restriction endonucleases) as a defense mechanism against foreign DNA. Just like other organisms, bacteria can be infected by viruses. However, these viruses are specific to bacteria and are called bacteriopahges. When a bacteriophage infects a bacterium, the bacteriophage injects its genetic material into the bacterium. In an attempt to defend itself, the restriction enzymes within the bacterium recognize specific DNA sequences that are unique to the viral DNA. Once the enzyme recognizes the viral DNA, it cuts them into pieces rendering harmless the bacterium.
How are restriction enzymes classified?
Restriction Enzymes are classified based on their activity sites, required cofactors, and recognition sequences. The detailed classification and description of each type of restriction endonucleases are presented in Table 1.
What temperature does a restriction enzyme digest?
The digestion activity of restriction enzymes depends on the following factors: Temperature: Most endonucleases digest the target DNA at 37 °C with few exceptions. Some work at lower temperatures (~25 °C, Sma 1) while Taq I works at 65 °C.
What is methylation in DNA?
Methylation of DNA is a major factor that affects the specificity of restriction endonucleases. DNA isolated from strains of bacteria expressing methylases such as Dam or Dcm may be resistant to cleavage by endonucleases as the recognition sites are methylated. Plasmid DNA isolated from E. coli expressing Dam is methylated at the GATC sites making it resistant to cleavage by Mbo I. Certain methylation-specific endonucleases also degrade methylated DNA without affecting the methylated host DNA. Bam H I isolated from Bacillus amyloliquefaciens H (Catalog Number R0260) cleaves methylated GGATCC sites of plasmid from E. coli expressing Dam. Dpn I (Catalog Number R8381) isolated from Diplococcus pneumonia targets methylated T7 DNA.
What cofactors are required for restriction endonuclease?
Cofactors: Restriction endonucleases require certain cofactors or combination of cofactors to digest at the recognition site. All enzymes require Mg 2+ as a cofactor for the endonuclease activity. In R-M systems with separate proteins having the restriction and methylation activities, S-adenosylmethionine (SAM) and ATP are required for methylation activity.
What is the function of restriction endonucleases?
The function of restriction endonucleases is mainly protection against foreign genetic material especially against bacteriophage DNA. The other functions attributed to these enzymes are recombination and transposition. Restriction endonucleases make up the restriction-modification (R-M) systems comprised of endonuclease ...
How to name endonuclease?
The guidelines for naming restriction enzymes are based on the original suggestion by Smith and Nathans. 1 The enzyme names begin with an italicized three-letter a cronym; the first letter of the acronym is the first letter of the genus of bacteria from which the enzyme was isolated, the next two letters are the two letters of the species. These are followed by extra letters or numbers to indicate the serotype or strain, a space, then a Roman numeral to indicate the chronology of identification. For example, the first endonuclease isolated from Escherichia coli, strain RY13 is named as Eco R I. Hin d III is the third endonuclease of four isolated from Haemophilus influenza, serotype d.
What is the sequence of DNA that is not methylated?
Each restriction enzyme recognizes a specific sequence of 4–8 nucleotides in DNA and cleaves at these sites. Endonucleases isolated by different organisms with identical recognition sites are termed isoschizomers.
What is a restriction enzyme?
A restriction enzyme is a DNA-cutting enzyme that recognizes specific sites in DNA. Many restriction enzymes make staggered cuts at or near their recognition sites, producing ends with a single-stranded overhang. If two DNA molecules have matching ends, they can be joined by the enzyme DNA ligase.
Where are restriction enzymes found?
Restriction enzymes are found in bacteria (and other prokaryotes). They recognize and bind to specific sequences of DNA, called restriction sites. Each restriction enzyme recognizes just one or a few restriction sites. When it finds its target sequence, a restriction enzyme will make a double-stranded cut in the DNA molecule.
What is the enzyme that seals the gap between two DNA molecules?
If two DNA molecules have matching ends, they can be joined by the enzyme DNA ligase. DNA ligase seals the gap between the molecules, forming a single piece of DNA. Restriction enzymes and DNA ligase are often used to insert genes and other pieces of DNA into plasmids during DNA cloning.
How is recombinant plasmid produced?
Right: recombinant plasmid produced when gene goes in backwards ("pointing" back towards the promoter that is already in the plasmid). Restriction digests and ligations like this one are performed using many copies of plasmid and gene DNA. In fact, billions of molecules of DNA are used in a single ligation!
Why are blunt-ended fragments harder to ligate together?
However, blunt-ended fragments are harder to ligate together (the ligation reaction is less efficient and more likely to fail) because there are no single-stranded overhangs to hold the DNA molecules in position.
How does DNA ligase work?
How does DNA ligase do this? Using ATP as an energy source, ligase catalyzes a reaction in which the phosphate group sticking off the 5’ end of one DNA strand is linked to the hydroxyl group sticking off the 3’ end of the other. This reaction produces an intact sugar-phosphate backbone.
Why do enzymes leave sticky ends?
Sticky ends are helpful in cloning because they hold two pieces of DNA together so they can be linked by DNA ligase. Not all restriction enzymes produce sticky ends.
Overview
Applications
Isolated restriction enzymes are used to manipulate DNA for different scientific applications.
They are used to assist insertion of genes into plasmid vectors during gene cloning and protein production experiments. For optimal use, plasmids that are commonly used for gene cloning are modified to include a short polylinker sequence (called the multiple cloning site, or MCS) rich in restriction enzyme recognition sequences. This allows flexibility when inserting gene fragments …
History
The term restriction enzyme originated from the studies of phage λ, a virus that infects bacteria, and the phenomenon of host-controlled restriction and modification of such bacterial phage or bacteriophage. The phenomenon was first identified in work done in the laboratories of Salvador Luria, Jean Weigle and Giuseppe Bertani in the early 1950s. It was found that, for a bacteriophage λ that can grow well in one strain of Escherichia coli, for example E. coli C, when grown in another …
Origins
Restriction enzymes likely evolved from a common ancestor and became widespread via horizontal gene transfer. In addition, there is mounting evidence that restriction endonucleases evolved as a selfish genetic element.
Recognition site
Restriction enzymes recognize a specific sequence of nucleotides and produce a double-stranded cut in the DNA. The recognition sequences can also be classified by the number of bases in its recognition site, usually between 4 and 8 bases, and the number of bases in the sequence will determine how often the site will appear by chance in any given genome, e.g., a 4-base pair sequence w…
Types
Naturally occurring restriction endonucleases are categorized into five groups (Types I, II, III, IV, and V) based on their composition and enzyme cofactor requirements, the nature of their target sequence, and the position of their DNA cleavage site relative to the target sequence. DNA sequence analysis of restriction enzymes however show great variations, indicating that there are more than four types. All types of enzymes recognize specific short DNA sequences and carry o…
Nomenclature
Since their discovery in the 1970s, many restriction enzymes have been identified; for example, more than 3500 different Type II restriction enzymes have been characterized. Each enzyme is named after the bacterium from which it was isolated, using a naming system based on bacterial genus, species and strain. For example, the name of the EcoRI restriction enzyme was derived as shown in the box.
See also
• BglII – a restriction enzyme
• EcoRI – a restriction enzyme
• HindIII – a restriction enzyme
• Homing endonuclease
What Are Restriction Enzymes?
- The restriction enzyme is a protein produced by bacteria that cleaves the DNA at specific sites. This site is known as the restriction site. The restriction enzymes protect the live bacteria from bacteriophages. They recognize and cleave at the restriction sites of the bacteriophage and destroy its DNA. Restriction enzymes are important tools for g...
Types of Restriction Enzymes
- Type I
These restriction enzymes cut the DNA far from the recognition sequences. However, they do not produce discrete restriction fragments, hence, are of not much practical value. These are complex, multi-subunit restriction and modification enzymes. They were initially thought to be ra… - Type II
These enzymes cut at specific positions closer to or within the restriction sites. Discrete restriction fragments and gel banding patterns are observed. They are exclusively used for DNA analysis and gene cloning in the laboratories. These are a family of unrelated proteins. They are …
Applications of Restriction Enzymes
- They are used in RFLP techniques to cut the DNA into smaller fragments to study the fragment length differences among the individuals.
You Might Also Be Interested in