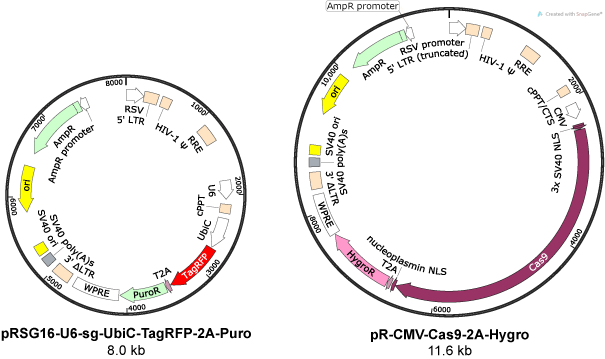
Creating a Plasmid from Scratch.
- Step 1: Importing the Plasmid Backbone from addgene into Benchling. What’s Benchling? It’s essentially the fullstack software of genetic engineering ...
- Step 2: Picking a suitable restriction site. We’d ideally pick a restriction site within a multiple cloning site, with 1 cut site. I chose Sac I ...
- Step 3: Inserting the TEF 1 promoter. These plasmids are often created in the lab, using an assembly procedure called the Gibson Assembly. It’s ...
- Step 4: Inserting the Kozak Sequence. The Kozak sequence (GCCGCCACCAUGG) functions as the translation initiation site in most eukaryotic mRNA ...
How to make a plasmid?
- Add 20 ml GMM with appropriate supplements and yeast extract at 5g/L into a petri dish.
- Inoculate with several loopful of Aspergillusconidia from solid media.
- Incubate at 37 °C for 16–24 hrs. ...
- Pour off media and scrape hyphal material together. ...
- Take cell material and place between paper towels and squeeze out remaining liquid. ...
What is the structure and function of a plasmid?
What is Plasmid?
- Plasmid Structure. Plasmids are extrachromosomal and not essential.
- Plasmid Vector. Plasmids and bacteriophages are frequently used as a cloning vector in the DNA recombinant technology.
- pBR322 Plasmid. Different antibiotic resistance genes act as a restriction site and to ligate foreign DNA and for the selection of transformants.
- Ti Plasmid. ...
How can a plasmid be desribed?
Plasmids are small, circular molecules of double-stranded DNA derived from larger plasmids that occur naturally in bacteria. 68 Most plasmid-cloning vectors are designed to replicate inE. coli. 69 All of the enzymes required for replication of the plasmid DNA are produced by a host bacterium. The classic example of plasmid vector is pBR322, which was one of the first such vectors to be recognized.
What is the process of the plasmid?
Plasmid or vector transformation is the process by which exogenous DNA is transferred into the host cell.Transformation usually implies uptake of DNA into bacterial, yeast or plant cells, while transfection is a term usually reserved for mammalian cells.

How do you design a plasmid?
2:4620:27Plasmid design (bacterial expression vector) - YouTubeYouTubeStart of suggested clipEnd of suggested clipBasically we cut the plasmid. Open. We also cut the ends of this pcr product with the same enzymes.MoreBasically we cut the plasmid. Open. We also cut the ends of this pcr product with the same enzymes. Then what we do is to we have to purify. Those we can't just go straight forward we have to purify.
How do you construct a plasmid cloning vector?
0:002:02Construction of a Plasmid Vector [HD Animation] - YouTubeYouTubeStart of suggested clipEnd of suggested clipPlasmids can be good cloning vectors because they carry an origin of replication. And are thereforeMorePlasmids can be good cloning vectors because they carry an origin of replication. And are therefore able to replicate independently. Within a cell. Most plasmids used as vectors also encode some type
How do you build a recombinant plasmid?
If DNA ligase was added to the tube of cut DNA, the sugar-phosphate backbone bonds would be sealed and the two cut linear DNA molecules would become one circular recombinant plasmid. This plasmid will have an unaltered Amp antibiotic resistance gene and an inactivated LacZ gene with the insertion.
How long does it take to construct a plasmid?
At the Broad Institute, Doench's lab has created hundreds of thousands of plasmids either en masse for genome wide screens or for the more traditional “one plasmid for one problem” scenario. He did the math and came to the conclusion that it takes 5 hours of hands-on time to make a plasmid.
How is a plasmid DNA template made?
DNA templates are synthesized by PCR using primers that start with a T7 or SP6 promoter sequence; linear PCR products are precipitated and resuspended in H2O to a concentration of 0.5–1 μg/μL.
What are the steps to making recombinant DNA?
0:278:16Steps in Recombinant DNA technology or rDNA ... - YouTubeYouTubeStart of suggested clipEnd of suggested clipTechnology step one it is a identification. And isolation of gene of interest or dna fragment to beMoreTechnology step one it is a identification. And isolation of gene of interest or dna fragment to be clause. Second we have to introduce this isolated gene into a suitable. Vector.
How do you make a recombinant DNA model?
3:365:21How to make a Recombinant DNA - YouTubeYouTubeStart of suggested clipEnd of suggested clipThe cut plasmids are mixed with the gene sized pieces of linear DNA in hope that they will bind withMoreThe cut plasmids are mixed with the gene sized pieces of linear DNA in hope that they will bind with each other with the help of their sticky. Ends.
How do we make recombinant DNA?
Recombinant DNA is the method of joining two or more DNA molecules to create a hybrid. The technology is made possible by two types of enzymes, restriction endonucleases and ligase. A restriction endonuclease recognizes a specific sequence of DNA and cuts within, or close to, that sequence.
What are the basic components of a plasmid vector?
components of plasmid cloning vectors:origin of replication (ori) site where DNA replication is initiated. ... marker genes for selection and/or screening. selection – killing cells that lack specific gene. ... Unique restriction endonuclease (RE) sites. ... transmissability. ... Promoters for gene expression.
What are 3 important parts of a plasmid vector?
Plasmids contain three components: an origin of replication, a polylinker to clone the gene of interest (called multiple cloning site where the restriction enzymes cleave), and an antibiotic resistance gene (selectable marker).
How many stop codons are there in an ORF?
The stop codon is usually included in the ORF otherwise there are often 2-3 shifted stop codons for the 3 frames downstream the polylinker therefore you're sure of it.
What is the origin of replication?
ORI is usually included in the plasmid, it is the origin of replication in order to make the plasmid replicates in the transformed host (prokaryotic) or transfected host (Eukaryotic), so there are ori for prokaryotes (example, OriE. to E.coli) or ori for other eukaryotic cells.
What is the inducible cloning?
To perform a cloning to expression use the inducible one, because usually you let grow transformed host and when you have a good amount of cells you induce expression with different compounds (related to the kind of promoter). Cloning: plasmid, host, gene product usage, etc....
Where is the stop codon in the ORF?
The stop codon is usually included in the ORF otherwise there are often 2-3 shifted stop codons for the 3 frames downstream the polylinker therefore you're sure of it. The promoter is usually upstream of the polylinker in the plasmid, to express the gene when you want (eg if it is a inducible one: lac trp etc ...) or constitutive ...
Why join Researchgate?
Join ResearchGate to find the people and research you need to help your work.
Do you have to insert a complete gene into a plasmid?
In this case, of course you have a complete gene to insert in a plasmid.
How is a plasmid constructed in the lab?
Due to their artificial nature, lab plasmids are commonly referred to as “vectors” or “constructs.” To insert a gene of interest into a vector, scientists may utilize one of a variety of cloning methods (restriction enzyme, ligation independent, Gateway, Gibson, etc). The cloning method is ultimately chosen based on the plasmid you want to clone into. Regardless, once the cloning steps are complete, the vector containing the newly inserted gene is transformed into bacterial cells and selectively grown on antibiotic plates.
How do scientists use plasmids?
Generally, scientists use plasmids to manipulate gene expression in target cells. Characteristics such as flexibility, versatility, safety, and cost-effectiveness enable molecular biologists to broadly utilize plasmids across a wide range of applications. Some common plasmid types include cloning plasmids, expression plasmids, gene knock-down plasmids, reporter plasmids, viral plasmids, and genome engineering plasmids.
What is the cloning method?
The cloning method is ultimately chosen based on the plasmid you want to clone into. Regardless, once the cloning steps are complete, the vector containing the newly inserted gene is transformed into bacterial cells and selectively grown on antibiotic plates.
What are plasmids used for?
Some of the many things that plasmids can be used to do include: 1 Produce large amounts of a protein so that scientists can purify and study it in a controlled setting. Read more:#N#Plasmids 101: Protein Tags 2 Produce proteins that glow so that scientists can track their location or quantity inside a cell#N#Plasmids 101: Green Fluorescent Protein (GFP)#N#Plasmids 101: Luciferase 3 Monitor the level of a chemical in a particular environment 4 Produce enzymes that will make specific, controlled changes to an organism’s genome ( genome engineering) 5 Produce synthetic viruses that can be used in research or for therapeutics
Why do scientists need to make plasmids?
Importantly, because the bacteria from which plasmids are isolated grow quickly and make more of the plasmids as they grow, scientists can easily make large amounts of plasmid to manipulate and use in later work.
What is the origin of plasmids?
All natural plasmids contain an origin of replication (which controls the host range and copy number of the plasmid) and typically include a gene that is advantageous for survival, such as an antibiotic resistance gene.
What is the function of Addgene?
Addgene has compiled various educational resources to facilitate plasmid use in the lab.
How much time does it take to make a plasmid?
This question sparked much debate around the office so we took to social media to see if we could come to a consensus. Though waiting is one of the greatest time sinks in molecular biology (or many other biological experiments, for that matter), we were interested in finding out the total hands-on time for cloning design, PCR, insertion in the backbone, transformation into bacteria, plasmid preps, and sequencing. This “total hands-on time” per plasmid assumes only one plasmid is being made at a time and that the correct clone reveals itself the first time.
How much time has Addgene saved?
We’re very excited to reach one million plasmids shared. Reagent sharing saves both time and money and based on Doench’s calculations, Addgene and the scientist who have deposited their plasmids at Addgene have saved the scientific community over $50 million dollars, at a generous minimum. In terms of total time Addgene has saved thus far, multiply 5 hours per plasmid by a million. Five million hours. According to Doench, "5 million hours, in case you were wondering, is the amount of time between the years 2018 and 1448, which is when the Inca were starting to build Machu Picchu, and Leonardo da Vinci hadn't been born yet." We’re proud to have such a positive impact on the scientific community and are eager to continue accelerating science through the next million plasmids and beyond!
How to save time when creating complex constructs?
While methods such as Gibson assembly can save you time when creating complicated constructs, you may also need to balance speed with cost. Create several constructs at once. Instead of making one plasmid at a time, try to group the construction of several plasmids in parallel. That way, you can run one PCR, one gel, etc. for all plasmids at once.
How long does it take for plasmids to ship?
Plasmids also ship within 2-3 business days which ensures you get the correct reagents quickly. Ask the PI or corresponding author of the published plasmids. This is a great way to network and build collaborations while obtaining the reagents you need. But unfortunately, this method does not have a 100% success rate and may require repeated ...
Who is John Doench?
At the same time, John Doench, an Addgene Blue Flame depositor, calculated the time to make a plasmid to help us settle the debate. At the Broad Institute, Doench’s lab has created hundreds of thousands of plasmids either en masse for genome wide screens or for the more traditional “one plasmid for one problem” scenario.
Can you add videos to your watch history?
Videos you watch may be added to the TV's watch history and influence TV recommendations. To avoid this, cancel and sign in to YouTube on your computer.
What sequence is used for cloning ORF?
Therefore, our Forward Primer will use the sequence 5'-ATGTGGCATATCTCGAAGTAC-3' for the region that binds the ORF and we will add the EcoRI restriction site (GAATTC) to the 5’ end of this primer, making our Forward Primer 5'-GAATTCATGTGGCATATCTCGAAGTAC-3'.
What are the primers used for cloning?
The basic PCR primers for molecular cloning consist of: 1 Leader Sequence: Extra base pairs on the 5' end of the primer assist with restriction enzyme digestion (usually 3-6bp) 2 Restriction Site: Your chosen restriction site for cloning (usually 6-8bp) 3 Hybridization Sequence: The region of the primer that binds to the sequence to be amplified (usually 18-21bp)
How long does it take to digest a plasmid?
We recommend using your entire PCR reaction and 1μg of recipient plasmid. It is also critical that as much of the recipient plasmid as possible be cut with both enzymes, and therefore it is important that the digest goes at least 4 hours and as long as overnight.
What is PCR cloning?
PCR based cloning is incredibly versatile and allows for nearly any piece of DNA to be placed into a backbone vector of choice with minimal limitations.
Why is it important to run a clone on a gel?
When cloning by PCR, it is especially important to run the product on a gel. This allows you to visualize that your PCR product is the anticipated size and that your band is strong ( indicating that the PCR reaction worked and that you have a sufficient amount of DNA).
What is the leader sequence?
Leader Sequence: Extra base pairs on the 5' end of the primer assist with restriction enzyme digestion (usually 3-6bp)
How to isolate insert and vector?
Isolate your insert and vector by gel purification: Run your digest DNA on an agarose gel and conduct a gel purification to isolate the DNA. When running a gel for purification purposes it is important to have nice crisp bands and to have space to cut out the bands.
How to denote a mutation in plasmid?
The proper way to denote an amino acid mutation is to list the one letter abbreviation of the wild type amino acid immediately followed by its position (number) relative to the start Methionine (Met) followed by the one letter abbreviation of the mutated amino acid currently at that position.
How many letters are in a plasmid name?
Include information about the insert in your plasmid name. This is often a 3-6 letter representation of the gene (or DNA sequence).
What is the first letter of a plasmid?
Tip: A lowercase "p" is often used as the first letter of a plasmid name and simply denotes that the object is a ‘plasmid’.
Where to list fusion proteins?
Typically you would list any tag or fusion protein in the order they appear in the plasmid and their relative position to the insert. Example, if you have a Flag tag on the N-terminal of your insert, you would list it first.
What Does “Ori” Mean?
“ Ori ” means the origin of plasmid replication. Whatever you do, don’t change it! Once a plasmid is unable to replicate, it is useless.
What is the ccdb gene in ptlnx?
The pTLNX vector also has a gene for plasmid selection (ccdB), along with virus SV40 nuclear localization signal and Xenopus globine 3’ UTR, that allows for high expression levels of cloned genes.
How many base pairs are in a plasmid?
As you see from the map center, the size of the linearized plasmid is 4361 base pairs. Before you start working with any plasmid, it is advisable to linearize it by cutting with a unique restriction enzyme to check that the advertised size is roughly the same as expected.
What are the two antibiotic resistance genes in pBR322?
pBR322 has two antibiotic resistance genes: tet (tetracycline resistance) and amp (ampicillin resistance). These genes encode an efflux pump ( tetR) and beta-lactamase ( ampR) to excrete tetracycline and ampicillin from the cell, respectively. Tet and amp are read in different directions.
What are the transcription promoters and terminators of plasmids?
In addition to genes, plasmids often include transcription promoters and terminators derived from E.coli phages. Promoters from phages SP6 and T7 are often used for in vitro RNA amplification. They require phage polymerases and are therefore inactive in vivo.
What is NEB in biology?
Established in the mid 1970's, New England Biolabs, Inc. (NEB) is the industry leader in the discovery and production of enzymes for molecular biology applications and now offers the largest selection of recombinant and native enzymes for genomic research. NEB continues to expand its product offerings into areas related to PCR, gene expression, ...
What are restriction sites?
Restriction sites for corresponding enzymes are shown as vertical lines with the position of the starting nucleotides. The sites should be unique, but it pays to check, as derivative vectors often contain additional forgotten sequences.
What happens to phage DNA during lytic phase?
In the course of lytic phase the phage DNA remains independent in the cell, replicates, codes for capsid proteins and forms large number of phage particles within the cell, result ing in the lysis (breakage) of the bacterial cell and thus releasing the phage particle.
What are the steps involved in the preparation of a recombinant DNA?
The steps are: 1. Selection of Target DNA 2. Selection of a Suitable Cloning Vector DNA or Vehicle DNA 3. Selection of Restriction Endonucleases 4.
How does phage infection occur?
The infection is caused by the phage particle by attacking to the outside of the bacterium and injecting its DNA chromosome into the cell. This DNA, about 50 kb, linear-double stranded, with single stranded complementary ends of 12 nucleotides in length (cohesive ends), gets arranged in circle in the host cell through pairing of cohesive ends and is transcribed as circular molecule, during early phase of infection. During this phase the phage may either adopt lytic phase or lysogenic phase.
How is DNA purified?
Then the DNA is purified from the cell extract for which the extract is treated with proteases and endonucleases and then the proteins precipitated with phenol and chloroform and finally centrifuged. The DNA will be measured in a spectrophotometer at 260 nm, at this wavelength the absorbance (A 260) of 1.0 corresponds to 50pg of double-stranded DNA/ml.
Why are cosmids used in genomics?
Due to their capacity to hold large sized DNAs, cosmids are used to construct genomic library i.e. a set of recombinant genes that contains the entire DNA present in an individual organism. The construction of libraries in bacteriophage X vectors has proven to be an effective means of isolating segments of DNA from complex eukaryotic genomes.
What is a cosmid?
Cosmid is a cloning vector consisting of the lambda cos site (single strand ed cohesive extensions at the ends of the DNA molecule) insert ed into a plasmid, used to clone DNA fragments upto 40 Kb in size.
What is a cloning vector?
The cloning vector is the DNA molecule into which the target DNA is introduced producing the recombinant DNA molecule. A good cloning vehicle is one which has only a single site for cutting by a particular restriction endonuclease. There are different types of vectors which can be used to clone fragments of foreign DNA and propagate (clone) ...

Plasmids
Origin of Replication
- An ORI is a specific sequence of DNA that initiates replication within a plasmid. It does this, by recruiting plasmid-encoded transcriptional proteins which recognise specific DNA sequences & determines it as the ORI. Origins of replication are typically either relaxed or stringent. Relaxed ORIs occur freely within the cytoplasm, and often yield high copy numbers of plasmids. Stringen…
Restriction Sites & Multiple Cloning Sites
- Restriction enzymes (endonucleases) are like killer robots, programmed with a single target in mind — to cleave a complementary 4–8p sequence, called a restriction site. They make a cut at that site, through two strands of DNA, typically leaving a 2–4 nucleotide single-stranded overhand in its wake. Oftentimes, within commercially-engineered plasmids, restriction sites are grouped t…
Gene Insert
- The gene of interest is what creates new functionality within the plasmid, and is the reason for the experiment in the first place. The gene of interest can be virtually anything, from GFP that causes cells to fluoresce in green to Bt which expresses insecticidal protein in its cells! After the same aforementioned restriction enzyme isolates the gene, it’s inserted into the plasmid using DNA lig…
Selection Markers
- Once the gene-of-interest has been inserted into the plasmid, it’s now used as a vector, to be inserted into a living cell! This often comes in the form of E. coli, a well-characterised & simple bacterial cell. The problem is, successful plasmid transformation (the uptake of the plasmid) is notoriously low, with only 1 out of 10, 000 cells succeeding on average. To quickly filter through …