Which codons are the stop signals?
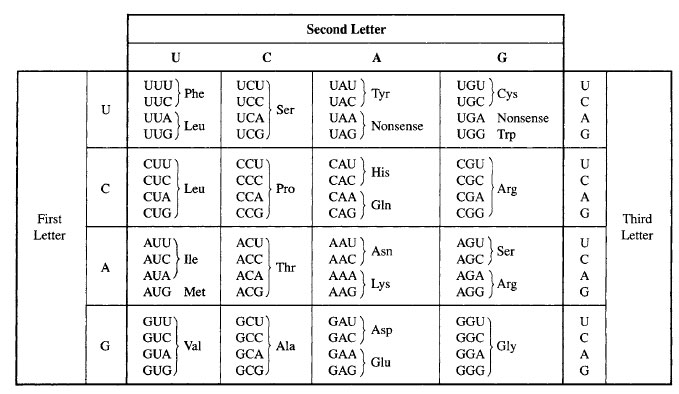
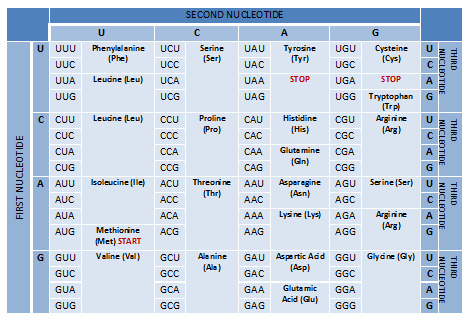
• Three codons (UAA, UAG, and UGA) are known as stop codons and they signal the termination of translation. They do not encode amino acids. • In special cases, the stop codons UGA and UAG can encode special 21st and 22nd amino acids. • One codon (AUG) is known as a start codon and initiates the process of translation.
What is the total possible number of codons?
• A total of 64 codons are possible. • The genetic code is universal because it is the same among all organisms. • Three codons, UAA, UAG, and UGA, are known as stop codons as they stop the process of translation. If this is a mutation as a stop codon and its finishes prematurely it is called an amber codon.
What is the codon sequence for the three stop codons?
There are 3 STOP codons in the genetic code - UAG, UAA, and UGA. These codons signal the end of the polypeptide chain during translation. These codons are also known as nonsense codons or termination codons as they do not code for an amino acid. The three STOP codons have been named as amber (UAG), opal or umber (UGA) and ochre (UAA).
What do start and stop codons do?
Start codon marks the site at which translation into protein sequence begins while stop codon marks the site at which translation ends. There are three stop codons as UAG, UAA, and UGA, and they do not code for an amino acid while the most common start codon AUG codes for methionine.
How many types of stop codons exist?
3 STOP codonsThere are 3 STOP codons in the genetic code - UAG, UAA, and UGA. These codons signal the end of the polypeptide chain during translation. These codons are also known as nonsense codons or termination codons as they do not code for an amino acid.
How many start codons are there?
The findings, to be published on February 21, 2017, in the journal Nucleic Acids Research by scientists in a research collaboration between NIST and Stanford University, demonstrate that there are at least 47 possible start codons, each of which can instruct a cell to begin protein synthesis.
How often is there a stop codon?
This is estimated to occur as often as 20% of the time. Thus a protein coding sequence downstream of a pseudoknot or a single stop codon will be synthesized at a lower quantity (~20%) than the protein encoded when the signal is ignored (~80% of the time).
What is the stop codon called?
Stop codons are also called nonsense codons because they do not code for an amino acid and instead signal the end of protein synthesis.
Why are there 3 stop codons?
Since codons are in no way separated, any synchronization shift during transcription or translation by ±n bases, where n is not divisible by three, produces a wrong sequence of triplets (see Fig. 1). Therefore, it seems very advantageous that nature invented three stop codons in the standard genetic code.
How do you remember stop codons?
The three stop codons are U A A, U G A and U A G. The mnemonic here is to think of an annoying person. When an annoying person comes up to you, you're gonna tell them: Stop, U Are Annoying, U Go Away, and U Are Gone. So that's the mnemonic for the three stop codons.
Is UAG a stop codon?
In the genetic code 11 (the bacterial, archaeal, and plant plastid code), the stop codons are UAA, UAG, and UGA. Frequency of stop codons in E. coli strain K-12 substr. W3110 was obtained from the codon usage database [3].
Is AUG a stop codon?
Each three-letter sequence of mRNA nucleotides corresponds to a specific amino acid, or to a stop codon. UGA, UAA, and UAG are stop codons. AUG is the codon for methionine, and is also the start codon.
Why is tag a stop codon?
Again, TAG, TAA, and TGA do not act as stop codons during transcription but are copied (substituting thymine for uracil) by RNA. Stop codons neither code for an amino acid nor belong to the non-coding group of genes but are a separate entity. Their recognition is much simpler than the recognition of the start codon.
Is TAA a stop codon?
1, B and C), TAA is the major stop codon. In the phyla Cyanobacteria and Proteobacteria, either TAA or TGA appears as the most frequent stop codon depending on the genomic GC content (Fig. 1, D and E). However, in all these phyla, TAG is the minor stop codon.
What are stop and start codons?
The start codon marks the site at which translation into protein sequence begins, and the stop codon marks the site at which translation ends.
Is ATG a start codon?
A start codon is made up of the letters ATG, which codes for the amino acid methionine. When the machinery of the cells sees that first ATG, it knows to start making the protein there.
What are the 3 start codons?
Each three-letter sequence of mRNA nucleotides corresponds to a specific amino acid, or to a stop codon. UGA, UAA, and UAG are stop codons. AUG is the codon for methionine, and is also the start codon.
Can you have two start codons?
In some cases, two ATG codons are closely located in the 5' end of mRNA, one might generate a truncated protein with few amino acid residues only, but another can result in a functional protein. In this case, the second one can be considered as start codon for that functional protein sequence.
What are the start and stop codons in DNA?
AUG, as the start codon, is in green and codes for methionine. The three stop codons are UAA, UAG, and UGA. Stop codons encode a release factor, rather than an amino acid, that causes translation to cease. Many scientists worked to decipher the genetic code.
What is the start codon in mRNA?
AUGThe start codon in all mRNA molecules has the sequence AUG and codes for methionine.
What is a stop codon?
In molecular biology (specifically protein biosynthesis ), a stop codon (or termination codon) is a codon ( nucleotide triplet within messenger RNA) that signals the termination of the translation process of the current protein. Most codons in messenger RNA correspond to the addition of an amino acid to a growing polypeptide chain, ...
How are stop codons distributed?
Distribution of stop codons within the genome of an organism is non-random and can correlate with GC-content. For example, the E. coli K-12 genome contains 2705 TAA (63%), 1257 TGA (29%), and 326 TAG (8%) stop codons (GC content 50.8%). Also the substrates for the stop codons release factor 1 or release factor 2 are strongly correlated to the abundance of stop codons. Large scale study of bacteria with a broad range of GC-contents shows that while the frequency of occurrence of TAA is negatively correlated to the GC-content and the frequency of occurrence of TGA is positively correlated to the GC-content, the frequency of occurrence of the TAG stop codon, which is often the minimally used stop codon in a genome, is not influenced by the GC-content.
What is the purpose of the codons in messenger RNA?
Most codons in messenger RNA correspond to the addition of an amino acid to a growing polypeptide chain, which may ultimately become a protein; stop codons signal the termination of this process by binding release factors, which cause the ribosomal subunits to disassociate, releasing the amino acid chain .
What is the stop codon in translation?
Stop codon suppression or translational readthrough occurs when in translation a stop codon is interpreted as a sense codon, that is, when a (standard) amino acid is 'encoded' by the stop codon. Mutated tRNAs can be the cause of readthrough, but also certain nucleotide motifs close to the stop codon. Translational readthrough is very common in viruses and bacteria, and has also been found as a gene regulatory principle in humans, yeasts, bacteria and drosophila. This kind of endogenous translational readthrough constitutes a variation of the genetic code, because a stop codon codes for an amino acid. In the case of human malate dehydrogenase, the stop codon is read through with a frequency of about 4%. The amino acid inserted at the stop codon depends on the identity of the stop codon itself: Gln, Tyr, and Lys have been found for the UAA and UAG codons, while Cys, Trp, and Arg for the UGA codon have been identified by mass spectrometry.
What are hidden stops?
Hidden stops are non-stop codons that would be read as stop codons if they were frameshifted +1 or −1. These prematurely terminate translation if the corresponding frame-shift (such as due to a ribosomal RNA slip) occurs before the hidden stop. It is hypothesised that this decreases resource waste on nonfunctional proteins and the production of potential cytotoxins. Researchers at Louisiana State University propose the ambush hypothesis, that hidden stops are selected for. Codons that can form hidden stops are used in genomes more frequently compared to synonymous codons that would otherwise code for the same amino acid. Unstable rRNA in an organism correlates with a higher frequency of hidden stops. This hypothesis however could not be validated with a larger data set.
How do nonstop mutations differ from nonsense mutations?
Nonstop mutations differ from nonsense mutations in that they do not create a stop codon but, instead, delete one. Nonstop mutations also differ from missense mutations, which are point mutations where a single nucleotide is changed to cause replacement by a different amino acid .
What is nonsense mutation?
Nonsense mutations are changes in DNA sequence that introduce a premature stop codon, causing any resulting protein to be abnormally shortened. This often causes a loss of function in the protein, as critical parts of the amino acid chain are no longer assembled. Because of this terminology, stop codons have also been referred to as nonsense codons .
What are the three STOP codons?
The three STOP codons have been named as amber (UAG), opal or umber (UGA) and ochre (UAA).
Why are the stop codons called ochre?
During protein synthesis, STOP codons cause the release of the new polypeptide chain from the ribosome. This occurs because there are no tRNAs with anticodons complementary to the STOP codons. Translation. Play.
What is the START codon for E. coli?
In prokaryotes, E. coli is found to use AUG 83%, GUG 14%, and UUG 3% as START codons. The lacA and lacI coding regions in the E coli lac operon don’t have AUG START codon and instead use UUG and GUG as initiation codons respectively.
What is the AUG codon?
START codons. The codon AUG is called the START codon as it the first codon in the transcribed mRNA that undergoes translation. AUG is the most common START codon and it codes for the amino acid methionine (Met) in eukaryotes and formyl methionine (fMet) in prokaryotes.
How many bases are in the universal genetic code?
The universal genetic code is made up of several codons or triplet bases. The standard code has evolved over time to minimize coding errors. There are a total of 64 codons in the genetic code arising from the permutation and combination of the 4 bases in nucleic acids.
What are the two punctuation marks in the genetic code?
There are two punctuation marks in the genetic code called the START and STOP codons which signal the end of protein synthesis in all organisms.
How many ways can you read DNA?
Each reading frame will produce a different sequence of amino acids and hence proteins. Thus, in double stranded DNA, there are 6 possible reading frames.
What is a stop codon?
Essentially, a stop codon is a specific cluster of nucleotides that tells protein construction mechanisms to stop chaining amino acids into a polypeptide chain. Stop codons are the bits of information that tell the body “Hey! This protein is finished!”. Stop codons work by initiating the release of release factors, ...
How do stop codons work?
Stop codons work by initiating the release of release factors, proteins that disassociate the ribosomal subunits and free the polypeptide chain. ADVERTISEMENT. In the human genetic code, there have been identified 3 stop codons, each represented as a triplet of nucleotide bases. In RNA: UAG (“amber”) UAA (“opal”)
How many amino acids are there in the human genome?
The exact order of amino acids determines the shape and function of the protein. All in all, there are 61 distinct codons in the human genome that encode for 20 amino acids. A table specifying the codons and the respective amino acids they signify. Credit: WikiCommons CC0 1.0.
What is the most common start codon in eukaryotic cells?
The most common start codon is the nucleotide triplet AUG (ATG in DNA). Almost every eukaryotic organism uses the triplet AUG as a start codon.
What are codons in mRNA?
So far, we have learned that codons are particular triplets of nucleotides that specify sequences of amino acids. The code in mRNA is read and a linear chain of amino acids is constructed according to the order of codons in the mRNA. Stop codons are special codons that tell the body to stop protein translation.
Why do the three stop codons appear differently in DNA and RNA?
The three stop codons appear differently in DNA and RNA because RNA contains the U base in place of the T base in DNA. ADVERTISEMENT. Stop codons are paired with “start codons” that tell the cellular machinery the beginning of a DNA sequence that specifies a specific protein.
Why do stop codons pop up?
Sometimes stop codons can pop up as the result of a random mutation. So-called “nonsense mutations” can introduce premature stop codons into the genome, which will cause early termination of translation. This is partially the reason why the DNA of so many organisms has evolved to have several redundant bits of code, so that if one fails others can take up the mantle.
How many stop codons are there in the human genome?
The start codon is only of use during the translation phase of protein synthesis and the same applies to the stop codon. Scientists agree that there are three stop codons – also called nonsense codons or termination codons – in the human genetic code.
What are the Three Stop Codons?
The three stop codons are TAG, TAA, and TGA in sense DNA, and UAG, UAA, and UGA in mRNA.
What is the start codon in DNA?
A DNA start codon usually carries the code ATG (in mRNA, this is AUG), although other codons have been discovered that also initiate the translation of genes such as GUG and UUG. The code AUG also produced an amino acid called methionine so other factors in the form of enzymes play a role in knowing when a start codon is, in fact, a start codon and not just another amino acid to add to the polypeptide chain. Messenger RNA does not understand the meaning of the start codon, whatever code this happens to be. It simply copies it. The start codon is only of use during the translation phase of protein synthesis and the same applies to the stop codon.
What are stop codons in protein synthesis?
Every organism’s DNA is constructed from sugar, phosphate, and the nucleotide bases cytosine (C), guanine (G), adenine (A), and thymine (T). DNA forms a double-helix structure of two connected strands; the nucleotide bases provide the connection points, each attaching to specific partners. Two identical bases cannot connect, such as thymine to thymine or adenine to adenine. These bases are also extremely limited as to their choice of partner. Thymine attaches to adenine, adenine to thymine, guanine bonds with cytosine, and cytosine with guanine. It is important to remember these fixed partners: TA/AT and CG/GC. Things get a little more complicated when portions of DNA code are copied onto a strand of RNA during the transcription process of protein synthesis. When this happens, uracil replaces thymine; the fixed partners in RNA are UA/AU and CG/GC.
How many codes does a nucleotide triplet have?
Leucine, serine, and arginine have six different codes per amino acid. This makes it less likely to produce the wrong protein in the presence of a genetic mutation. As the strand of mRNA makes its way through the ribosome, each amino acid links to the next to form a polypeptide chain (protein). The sequence of amino acids determines the type of protein, just as the sequence of nucleotide bases determines the type and order of the amino acids.
What is the function of stop codons?
A stop codon is a genetic code that signals the end of protein manufacturing inside the cell, like a period at the end of a sentence. The three stop codons are nucleotide base triplets that play an important role in intracellular protein synthesis; physiological and/or anatomical changes are possible if a stop codon is in the wrong position on a DNA or RNA strand, or if the code sequence is changed.
What is the code of RNA?
RNA then transcribes (copies) the code of the antisense strand in an antiparallel fashion, so from left to right – exactly like the sense strand. That means the code of the RNA will be AAAGCC – exactly the same as the code of the antisense strand of DNA.
What is a codon?
Codon. Codon. =. A codon is a trinucleotide sequence of DNA or RNA that corresponds to a specific amino acid. The genetic code describes the relationship between the sequence of DNA bases (A, C, G, and T) in a gene and the corresponding protein sequence that it encodes. The cell reads the sequence of the gene in groups of three bases.
What is the name of the sequence of nucleotides that are a specific amino acid?
Codon is the name we give a stretch of the three nucleotides, you know, one of A, C, G, or T, three of which in a row, that code for a specific amino acid, and so the genetic code is made up of units called codons where you have three nucleotides that code for a specific amino acid next to another three nucleotides, another three nucleotides, and another three nucleotides. And the cellular machinery, again the ribosome, that comes through and reads that genetic code, plugs in the correct amino acid that corresponds to each of the triplet code that's in the codon.
Is heterozygous genotype incomplete dominance?
The heterozygous genotype in an example of incomplete dominance has
Is BC FMET cleaved?
no, bc fmet and met are often cleaved from the amino acid chain after translation.

Overview
Properties
In the standard genetic code, there are three different termination codons:
There are variations on the standard genetic code, and alternative stop codons have been found in the mitochondrial genomes of vertebrates, Scenedesmus obliquus, and Thraustochytrium.
The nuclear genetic code is flexible as illustrated by variant genetic codes that reassign standard stop codons to amino acids.
Nomenclature
Stop codons were historically given many different names, as they each corresponded to a distinct class of mutants that all behaved in a similar manner. These mutants were first isolated within bacteriophages (T4 and lambda), viruses that infect the bacteria Escherichia coli. Mutations in viral genes weakened their infectious ability, sometimes creating viruses that were able to infect and grow within only certain varieties of E. coli.
Mutations
Nonsense mutations are changes in DNA sequence that introduce a premature stop codon, causing any resulting protein to be abnormally shortened. This often causes a loss of function in the protein, as critical parts of the amino acid chain are no longer assembled. Because of this terminology, stop codons have also been referred to as nonsense codons.
A nonstop mutation is a point mutation that occurs within a stop codon. Nonstop mutations caus…
Hidden stops
Hidden stops are non-stop codons that would be read as stop codons if they were frameshifted +1 or −1. These prematurely terminate translation if the corresponding frame-shift (such as due to a ribosomal RNA slip) occurs before the hidden stop. It is hypothesised that this decreases resource wastage on nonfunctional proteins and the production of potential cytotoxins. Researchers at Louisiana …
Translational readthrough
Stop codon suppression or translational readthrough occurs when in translation a stop codon is interpreted as a sense codon, that is, when a (standard) amino acid is 'encoded' by the stop codon. Mutated tRNAs can be the cause of readthrough, but also certain nucleotide motifs close to the stop codon. Translational readthrough is very common in viruses and bacteria, and has also been found as a gene regulatory principle in humans, yeasts, bacteria and drosophila. This kind …
Use as a watermark
In 2010 when Craig Venter unveiled the first fully functioning, reproducing cell controlled by synthetic DNA he described how his team used frequent stop codons to create watermarks in RNA and DNA to help confirm the results were indeed synthetic (and not contaminated or otherwise), using it to encode authors' names and website addresses.
See also
• Genetic code
• Start codon
• Terminator gene
Reading Frames
Start Codons
- The codon AUG is called the START codon as it the first codon in the transcribed mRNA that undergoes translation. AUG is the most common START codon and it codes for the amino acid methionine (Met) in eukaryotes and formyl methionine (fMet) in prokaryotes. During protein synthesis, the tRNA recognizes the START codon AUG with the help of some initiation factors a…
Stop Codons
- There are 3 STOP codons in the genetic code - UAG, UAA, and UGA. These codons signal the end of the polypeptide chain during translation. These codons are also known as nonsense codons or termination codons as they do not code for an amino acid. The three STOP codons have been named as amber (UAG), opal or umber (UGA) and ochre (UAA). "Amber" or UA...
References
Further Reading