What is the largest component of the human genome derived from?
00:28:06.12 It turns out that the largest component of genomes are derived from transposable elements. 00:28:18.19 more or less fifty percent are derived from transposable element sequences.
How are transposable elements changing the size of the human genome?
00:12:14.12 transposable elements are inserting massively between the genes and expanding the genome. 00:12:21.09 So this is largely responsible for the difference in genome size.
What are transposable elements and how are they formed?
Transposable elements come in many different forms and shapes. Transposable elements (TEs) are DNA sequences that have the ability to change their position within a genome. As a result of their deep evolutionary origins and continuous diversification, TEs come in a bewildering variety of forms and shapes (Fig. 1 ).
Which organisms have transposable elements in their genomes?
00:28:23.24 Plants, especially flowering plants have even higher proportions of their genomes that are transposable elements. 00:28:31.19 The maize genome, over 75% of the genome is derived from transposable elements. 00:28:37.16 The barley genome, and we will be talking more about these creatures in the next talk.

What are transposable elements?
Transposable elements (TEs) are DNA sequences that can alter their location in the genome. They are ubiquitous: ~ 48% of the human genome can be directly annotated as TE-derived [1]. TEs are of particular importance because they can modify or create genes and gene families [2–4]. Moreover, insertion of new TE copies into the genome frequently imposes a fitness cost, which results in an evolutionary arms race between active TE families and host factors that evolve to silence them [5, 6]. TEs have also been shown to modulate the expression nearby genes by acting as cis-regulatory elements (promoters, enhancers or repressors) [7–11]. In addition, they can contribute to numerous diseases through insertional mutagenesis by disrupting coding sequences or splicing [12–15] and developmental disorders [16]. Finally, there is substantial evidence that somatic TE insertion can upregulate oncogenes and cause genomic rearrangements to drive diverse cancers [17–20].
How many recently mobile transposable elements are there?
By using a novel statistical test we have defined a catalogue of 20 recently mobile transposable element subfamilies. We illustrate the gene regulatory potential of RMS-derived polymorphic TE insertions, using CRISPR/Cas9 deletion in vitro on a specific candidate, as well as by genome wide analysis of allele-specific expression. Our study presents novel insights into TE mobility and regulatory potential and provides a key resource for human disease genetics and population history studies.
Where is the 1Laboratory of Epigenomics and Chromatin Organization?
1Laboratory of Epigenomics and Chromatin Organization, Genome Institute of Singapore, A*STAR, Singapore, 138672 Singapore
Can HERV repeats be transposable?
It has been suggested that HERV repeats, which are endogenous retroviruses in the human genome, are no longer capable of transposition [54, 55]. However, an intact HERV-K provirus has been identified in a single individual with the potential for retained infectivity and a small number of HERV-K loci have shown evidence of polymorphic insertion [56, 57]. Although our analysis did not detect HERV-K per se, it did flag LTR5_Hs, the solo LTR created by recombination between the two near-identical LTR sequences at the flanks of HERV-Krepeats, as a highly significant RMS (FDR Q-value = 1.6e-16; Table Table1).1). Thus, our results provide statistical support to previous anecdotal reports that HERV-K has indeed been recently mobile.
What is the human genome made of?
Much of the human genome sequence is made up of interspersed repeats derived from the activity of transposable elements (TEs) 1, 2, mainly retrotransposons. Expression of retrotransposition-competent TEs can lead to more insertions. Therefore, limiting TE expression is fundamental to genome integrity, as the insertion of new elements into the human genome can disrupt gene function or alter gene expression, contributing to disease.
What is transposable element?
In numerous instances, transposable elements (TEs) have provided regulatory sequences and protein-coding domains that have been co-opted to serve the purposes of the host.
What is retrotransposition in genetics?
Retrotransposition events can result in large sequence insertions. This process creates a distinct ‘brand’ of genetic variation that is qualitatively different from nucleotide substitutions and other mutagenic mechanisms. That is, whereas mobile element insertion variants can be likened to copy number variants (CNVs) with respect to size, TE insertions differ qualitatively from the typical CNV as a result of their sequence content. Although CNVs can contain regulatory or coding sequences, their sequence content is often random. By contrast, TE insertions often encompass functional sequences intrinsic to the mobile element. Depending on the type of TE, an insertion polymorphism can include a promoter for autonomous expression, other regulatory sequences that lead to heterochromatin formation, marked secondary RNA or DNA structures and splicing regulators or protein-coding sequences. Another major difference between a TE insertion and a large-scale duplication is the copy number; thousands of TE copies exist in the human genome, which provide ample templates for pairing of these sequences in DNA, resulting in recombination and further genomic rearrangements, or in RNA, leading to altered splicing, editing or degradation.
How many copies of L1 are there in the human genome?
Similarly, L1 sequences are substantial constituents of other primate genomes 21. Although there are hundreds of thousands of copies of L1 in the human genome, only ~100 are fully intact, and a handful are highly active or ‘hot’ L1 (refs 22, 23 ).
Can TE be transcribed?
Both polymorphic alleles and fixed insertions can be autonomously transcribed. Expression of the so-called ‘unit’ or ‘authentic’ transcript from a TE locus 47, 48 — initiated from the TE promoter and encompassing primarily its sequence — is tightly regulated as a guard against retrotransposition and subsequent disruption of the genome, which could lead to disease. Most elements are silenced in normal differentiated somatic cells by heterochromatinization and DNA methylation 49, 50, 51, 52, 53, 54. Thus, TEs are most likely to be expressed as part of transcriptional ‘readthrough’ in the form of intronic sequences and lncRNAs 48. Because of their high copy number in the genome, definitively associating TE-encoded RNAs with specific loci is challenging using short-read sequencing, and it is a common pitfall to assume that expression of a TE subfamily reflects genome-wide transcription of retrotransposition intermediates (Box 2 ). When TE-encoded promoters truly escape silencing mechanisms, they can produce RNAs analogous to retrotransposition intermediates, TE-encoded proteins and chimeric transcripts that splice into nearby genes 46. TE-derived promoters can also regulate genes and initiate lncRNAs 55.
Is transposable element a part of the human genome?
Transposable elements are abundant in the human genome, and great strides have been made in pinpointing variations in these repetitive sequences using whole-genome sequencing. Now, the focus is shifting to understanding their expression and regulation, and the functional consequences of their insertion and retention in the genome over time.
What are transposable elements?
Transposable elements (TEs) are DNA sequences that have the ability to change their position within a genome. As a result of their deep evolutionary origins and continuous diversification, TEs come in a bewildering variety of forms and shapes (Fig. 1 ). TEs can be divided into two major classes based on their mechanism of transposition, and each class can be subdivided into subclasses based on the mechanism of chromosomal integration. Class 1 elements, also known as retrotransposons, mobilize through a ‘copy-and-paste’ mechanism whereby a RNA intermediate is reverse-transcribed into a cDNA copy that is integrated elsewhere in the genome [ 1 ]. For long terminal repeat (LTR) retrotransposons, integration occurs by means of a cleavage and strand-transfer reaction catalyzed by an integrase much like retroviruses [ 2 ]. For non-LTR retrotransposons, which include both long and short interspersed nuclear elements (LINEs and SINEs), chromosomal integration is coupled to the reverse transcription through a process referred to as target-primed reverse transcription [ 3 ]. Class 2 elements, also known as DNA transposons, are mobilized via a DNA intermediate, either directly through a ‘cut-and-paste’ mechanism [ 4, 5] or, in the case of Helitrons, a ‘peel-and-paste’ replicative mechanism involving a circular DNA intermediate [ 6 ]. For detailed reviews on individual TE types and transposition mechanisms, we refer the reader to the monograph edited by Craig et al. [ 7 ].
What is a TE in a genome?
TEs occupy a substantial portion of the genome of a species, including a large fraction of the DNA unique to that species . In maize, where Barbara McClintock did her seminal work [ 28 ], an astonishing 60 to 70% of the genome is comprised of LTR retrotransposons, many of which are unique to this species or its close wild relatives, but the less prevalent DNA transposons are currently the most active and mutagenic [ 29, 30, 31, 32] (Fig. 2 ). Similarly, the vast majority of TE insertions in Drosophila melanogaster are absent at the orthologous site in its closest relative D. simulans (and vice versa), and most are not fixed in the population [ 33, 34 ]. Many TE families are still actively transposing and the process is highly mutagenic; more than half of all known phenotypic mutants of D. melanogaster isolated in the laboratory are caused by spontaneous insertions of a wide variety of TEs [ 35 ]. Transposition events are also common and mutagenic in laboratory mice, where ongoing activity of several families of LTR elements are responsible for 10–15% of all inherited mutant phenotypes [ 36 ]. This contribution of TEs to genetic diversity may be underestimated, as TEs can be more active when organisms are under stress, such as in their natural environment [ 37, 38 ].
Why are TEs ignored in genomics?
TEs have been historically neglected and remain frequently ignored in genomic studies in part because of their repetitive nature, which poses a number of analytical challenges and often requires the use of specialized tools [ 187 ]. As genomes can harbor thousands of copies of very similar TE sequences, uniqueness or, alternatively, repetitiveness of substrings within these regions need to be taken into consideration during both experimental design and analysis. As an example, short DNA oligos targeting a specific TE instance in the genome for PCR, short hairpin RNA, or CRISPR-Cas9 have to be carefully designed and validated to ensure that they are truly specific and target unique regions of the genome. In some scenarios, it can be acceptable or even desirable to target many elements simultaneously [ 150] or an entire TE family [ 153, 188, 189, 190, 191 ].
How does transposition affect the evolution of the genome?
Transposition represents a potent mechanism of genome expansion that over time is counteracted by the removal of DNA via deletion. The balance between the two processes is a major driver in the evolution of genome size in eukaryotes [ 21, 50, 51 ]. Several studies have demonstrated the impact and range of this shuffling and cycling of genomic content on the evolution of plant and animal genomes [ 52, 53, 54, 55 ]. Because the insertion and removal of TEs is often imprecise, these processes can indirectly affect surrounding host sequences. Some of these events occur at high enough frequency to result in vast amounts of duplication and reshuffling of host sequences, including genes and regulatory sequences. For example, a single group of DNA transposons (MULEs) has been responsible for the capture and reshuffling of ~ 1,000 gene fragments in the rice genome [ 56 ]. Such studies have led to the conclusion that the rate at which TEs transpose, which is in part under host control, is an important driver of genome evolution [ 57, 58, 59 ].
Why are TE proteins important in evolution?
Thus, the biochemical activities of TE-derived proteins have been repeatedly co-opted during evolution to foster the emergence of convergent cellular innovations in different organisms. TEs can donate their own genes to the host, but they can also add exons and rearrange and duplicate existing host genes.
Where is reverse transcription of R2Bm RNA primed?
Luan DD, Korman MH, Jakubczak JL, Eickbush TH. Reverse transcription of R2Bm RNA is primed by a nick at the chromosomal target site: a mechanism for non-LTR retrotransposition. Cell. 1993;72:595–605.
Is transposition a dead end?
Historically, little attention has been given to transposition in somatic cells and its consequences, because somatic transposition may be viewed as an evolutionary dead-end for the TE with no long-term consequences for the host species. Yet, there is abundant evidence that TEs are active in somatic cells in many organisms [ 94] (Fig. 2 ). In humans, L1 expression and transposition have been detected in a variety of somatic contexts, including early embryos and certain stem cells [ 95, 96 ]. There is also a great deal of interest in mobile element expression and activity in the mammalian brain, where L1 transposition has been proposed to diversify neuronal cell populations [ 97, 98, 99 ]. One challenge for assessing somatic activity has rested with the development of reliable single cell insertion site mapping strategies [ 100, 101, 102, 103 ].
What are transposable elements?
Transposable elements (both active and inactive) occupy approximately half the human genome and a substantially greater fraction of some plant genomes! These movable elements are ubiquitous in the biosphere, and are highly successful in propagating themselves. We now realize that some transposable elements are also viruses, for instance, ...
What are the characteristics of transposable elements?
Beyond the common property of mobility, transposable elements show considerable diversity. Some move by DNA intermediates, and others move by RNA intermediates. Much of the mechanism of transposition is distinctive for these two classes, but all transposable elements effectively insert at staggered breaks in chromosomes. Some transposable elements move in a replicative manner, whereas others are nonreplicative, i.e. they move without making a copy of themselves.
How does transposition affect DNA?
In this manner, transposition can move DNA sequences that are not normally part of a transposable element to new locations. Indeed, "host" sequences can be acquired by viruses and propagated by infection of other individuals. This may be a natural means for evolving new strains of viruses. One of the most striking examples is the acquisition and modification of a proto-oncogene, such as cellular c-src, by a retrovirus to generate a modified, transforming form of the gene, called v-src. These and related observations provided insights into the progression of events that turn a normal cell into a cancerous one. They also point to the continual acquisition (and possibly deletion) of information from host genomes as a natural part of the evolution of viruses.
What is transposition related to?
Transposition is related to replication, recombination and repair. The process of moving from one place to another involves a type of recombination, insertions of transposable elements can cause mutations, and some transpositions are replicative, generating a new copy while leaving the old copy intact.
What is the mechanism of transposition?
Much of the mechanism of transposition is distinctive for these two classes, but all transposable elements effectively insert at staggered breaks in chromosomes. Some transposable elements move in a replicative manner, whereas others are nonreplicative, i.e. they move without making a copy of themselves.
What happens if a target gene is not expressed in a cell type?
If the target gene is not usually expressed in a certain cell type, this activation can lead to pathology, such as activation of a proto-oncogene causing a cell to become cancerous. In other cases, no obvious phenotype results from the transposition.
Can transposable elements activate nearby genes?
A particular type of transposable element can activate, inactivate or have no effect on nearby genes, depending on exactly where it inserts, it’s orientation and other factors. Figure 9.1. Possible effects of movement of a transposable element in the function and expression of the target gene.
How much of the human genome is retrotransposon?
Retrotransposon sequences comprise more than 40 % of the human genome [1,2]. Once dismissed as "junk DNA" of little or no adaptive significance [3,4], retrotransposons and other classes of transposable elements (TEs) are now generally considered as significant contributors to gene and genome evolution [5-9]. Of particular interest has been the ability of TEs to contribute to exon evolution by "exonization", i.e., an insertion of a TE into an intron and subsequent recruitment of this sequence or its part into a new protein-coding exon [10]. For example, it has been estimated that 5% of all alternatively spliced human exons had been derived from the exonization of Alu elements [11-13].
What are LTR transposable elements?
LTR transposable elements comprise nearly one-tenth of the human genome and have been implicated in the cis-regulatory evolution of a number of human genes [5,6,14-18]. The structure of a complete LTR retrotransposon (autonomous mobile element) comprises two copies of long terminal directed repeats (LTRs) flanking an internal region containing gag and pol genes, which encode a protease, reverse transcriptase, RNase H and integrase. These protein products are necessary for the formation of virus-like particles (VLPs) wherein replication of the element takes place. Some elements evolved from retroviruses have additional open reading frames (ORFs), e.g. env gene [1,19]. Flanking LTRs contain all the necessary transcriptional regulatory elements.
How did LRTS play a role in human gene evolution?
Our analysis of LRTS exonization events has shown that the patterns of LRTS distribution in human exons support the hypothesis that LRTS played a significant role in human gene evolution by providing cis-regulatory sequences; direct incorporation of LTR sequences into protein coding regions was observed less frequently. Combination of computational and experimental approaches used for tracing the history of the LTR exonization process of IL22RA2 gene presents a promising strategy that could facilitate further studies of transposon initiated gene evolution.
How many genes are associated with LTR?
We found that Long Terminal Repeat (LTR) retrotransposons are associated with 1,057 human genes (5.8%). In 256 cases LTR retrotransposons were observed in protein-coding regions, while 50 distinct protein coding exons in 45 genes were comprised exclusively of LTR RetroTransposon Sequence (LRTS). We go on to reconstruct the evolutionary history of an alternatively spliced exon of the Interleukin 22 receptor, alpha 2 gene (IL22RA2) derived from a sequence of retrotransposon of the Mammalian apparent LTR retrotransposons (MaLR) family. Sequencing and analysis of the homologous regions of genomes of several primates indicate that the LTR retrotransposon was inserted into the IL22RA2 gene at least prior to the divergence of Apes and Old World monkeys from a common ancestor (~25 MYA). We hypothesize that the recruitment of the part of LTR as a novel exon in great ape species occurred prior to the divergence of orangutans and humans from a common ancestor (~14 MYA) as a result of a single mutation in the proto-splice site.
How many genes have LRTS?
We further analyzed the abundance of LRTS-derived exons in gene transcripts. Most of the 275 genes containing at least one exon completely derived from LRTS (201 out of 275 ) are single transcript genes while the remaining 74 generate more than one transcript per gene. Note that about 60% (121/201) of single transcript genes encode zinc finger proteins (25%) or hypothetical proteins (35%). Apparently for the single transcript gene the LRTS insertion either has not disrupted the host gene function or possibly provided some beneficial modulation of the initial function and thus has been tolerated by natural selection.
Where do most of the protein coding exons come from?
Interestingly, most of the protein coding exons derived entirely from the LTR flanking regions originated from the MaLR family (24 out of 36). This could be explained by several factors. First of all, MaLR elements make up about 50% of the LTR retroelements in the human genome [1], and this high frequency alone may relate to their overrepresentation in protein coding exons. MaLRs are also relatively ancient elements, which have probably been exposed to more opportunities for exonizations over time. Note that the age factor has been implicated for proliferation of Alu-derived exons as well [12]. Finally, it is a formal possibility that nucleotide sequences of the MaLR family are better amenable for derivation of protein coding exons.
How many exons overlap with LTR?
We found that human gene exons (either protein-coding or non-coding) overlap with LTR flanks of LTR elements more frequently (1,074 cases) than with internal sequences (242 cases; note that exons overlaped with both regions were counted twice). This observation could be related to the fact that most (85%) of the LTR retroposon-derived sequences in human genome consist only of a solo LTR, with the internal sequence lost due to homologous recombination between the flanking LTRs [1]. Upon checking by BLASTX of 242 exons overlapping with the internal sequences, 61 exons were found to contain a section or even a whole viral gene (i.e. gag, pol, and env). However, only 20 of these 61 exons were protein-coding exons. Moreover, only in 10 cases was the reading frame of a human gene the same as the one of the viral gene. Seven out of these ten cases were observed in hypothetical genes. The remaining three cases represented a gene for endogenous retroviral protein, syncytin (ERVWE1), a gene for Krueppel-related zinc finger protein(H-plk) and a placenta-specific gene (PLAC4) which protein products contain the envelope, envelope and gag viral protein domain, respectively. All three genes are preferentially expressed in the placenta [23-25]. This observation indicates that the invasion of the Human Endogenous Retrovirus (HERV) may contribute to molecular mechanisms involved in human reproduction [26].
How much of the genome is made up of TEs?
Amazingly, as much as 50% of a mammalian genome and much more of a plant genome can be made of TEs. In Part 2 of her talk, Wessler discusses work from her lab analyzing the impact of TEs on gene and genome evolution. By looking for and finding a TE currently undergoing rapid amplification, Wessler and her colleagues have been able to assess how a type of TE called a MITE can rapidly increase its copy number without killing its host, rice.
When were transposable elements discovered?
00:09:56.08 In the 1950s transposable elements were discovered in Drosophila fruit fly. 00:10:01.15 In the 1960s, they were discovered in bacteria, in E. coli. 00:10:05.11 And in the 1970s they were discovered in the human genome.
Can 00:01:25.22 increase the copy number in the genome without harming the host significantly?
00:01:25.22 are able to increase their copy number in the genome without harming the host significantly.
Is genetic analysis limited?
00:03:09.15 Unfortunately the genetic analysis is limited in its scope.
What are transposable elements?
One group thinks that transposable elements have no function; rather, these are a type of “genetic parasite” that can spread within the host genome. These can be transmitted to the next generations as long as they do not have important adverse effects on the ability of the host to survive and reproduce. The other group argues that transposons are key element to cause genomic changes that fuel the biological evolution.
How long has the Alu family been in the human genome?
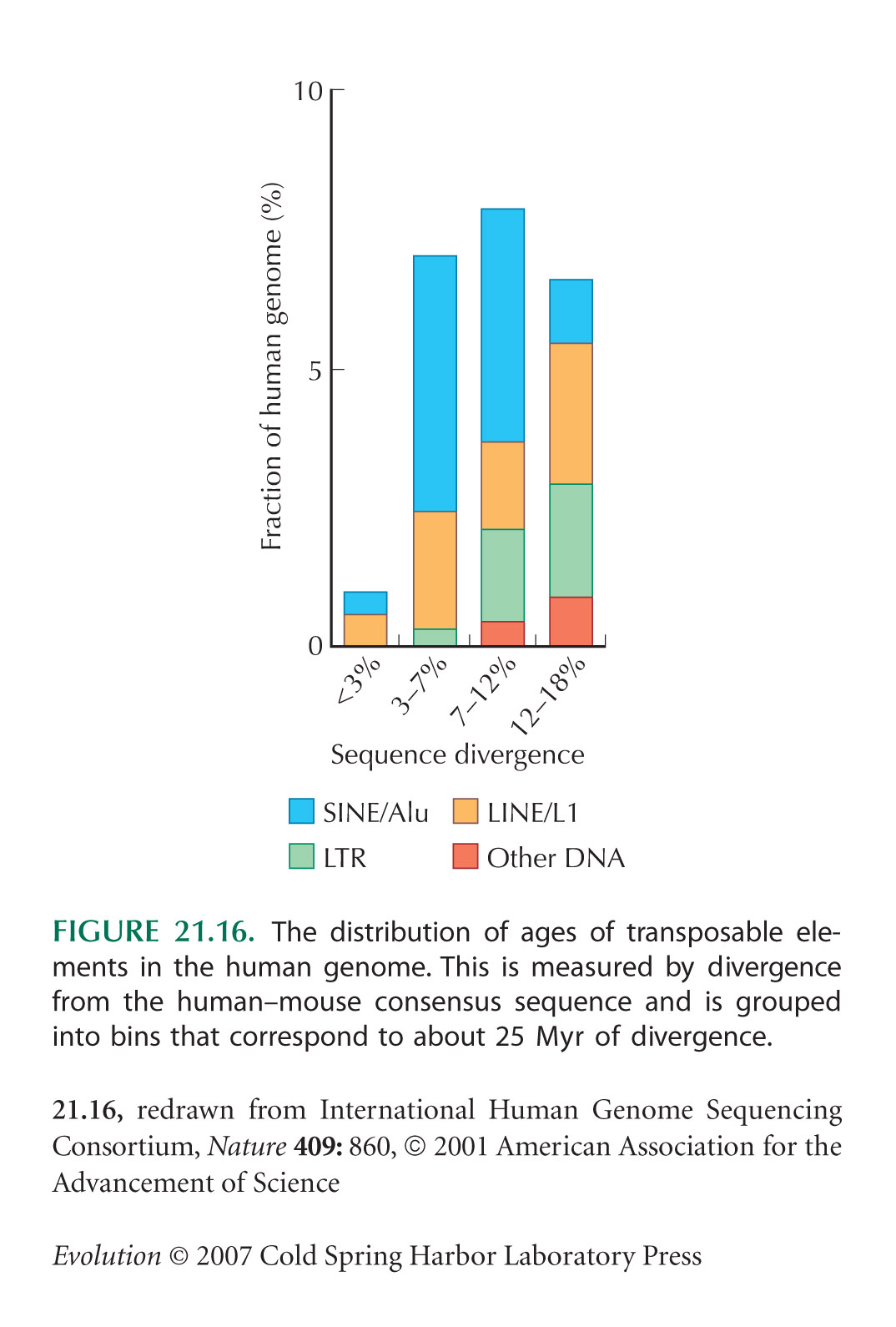
It was found that the average Alu family member has been in the genome of human evolutionary line for about 25 million years. This sequence does not repeat in genomes throughout the animal kingdom. However, whether its amplification is reaching a saturation point in these genome or is continuing unimpeded, cannot be determined with the present day data.
What is the enzyme that transposes DNA?
It ampunts up to 3 percent of the total genomic DNA. It contains the enzyme Alu 1 (A1 you one). The Alu sequence can transpose as evidenced from the study with patients of neurofibromatosis. DNA analysis of these patients showed that an Alu sequence was present in one of the introns of the neurofibromatosis gene.
What is the effect of the Alu sequence in the intron?
The RNA transcript of this gene was longer than those from normal individuals. The Alu sequence present in the intron disrupted the processing of the transcript, causing one exon to be lost completely from the mature mRNA. As a result the protein coded was 800 amino acids shorter than normal and was nonfunctional.
What are the two retrotransposons?
In human genome, two retro-transposons called LINEs (long interspersed sequences) and SINEs (short interspersed sequences) are found. LINEs are repeated sequences more than 5,000 bp long, interspersed among unique sequence DNA up to approximately 35,000 bp long. Full length LINEs are autonomous elements that encode the enzymes for their own retro-transposition and enzymes required for the transposition of SINEs.
How long has P been in the fruit fly genome?
Thus, it is thought that P elements have been introduced into the genome of a single fruit fly within the past fifty years or so, probably via transposition from a fly of another species and has rapidly spread through the entire fruit fly population.
When did Alu appear in the genome?
Studies of the genomes of various mammals indicate that the Alu sequence first appeared as a transposable element in the genome of higher primates about 60 million years ago. Ever since, it has been increasing in copy number at the rate of about one copy every 100 years.
