
One of the most important uses of genome science is as a tool to solve the medical riddles of disease. Scientists have identified specific locations within the map of the human genome where we think there are likely to be genes for key diseases like cancer, heart disease, or diabetes.
What does the genome consist of?
The genome is the entire set of genetic instructions found in a cell. In humans, the genome consists of 23 pairs of chromosomes, found in the nucleus, as well as a small chromosome found in the cells' mitochondria. Each set of 23 chromosomes contains approximately 3.1 billion bases of DNA sequence. Illustration Narration "Genome" is a funny word.
How big is a genome?
Your genome is broken into 23 chromosomes. In most cells there are two copies of each chromosome. If we take all of the DNA in a single nucleus, untangle it and stretch it out end-to-end, the entire human genome is about 2 meters (6.5 feet) long!!!
Why is the Human Genome Project so important?
What are the benefits of sequencing the human genome quizlet?
- one of the benefits of sequencing the human genome is that it will make it easier to identify genes that cause disease when they are. …
- What is the goal of physical mapping? …
- Cytogenetic mapping. …
- Linkage mapping. …
- Physical mapping.
Who is the guardian of the genome?
Dubbed the “guardian of the genome,” the p53 protein (blue) senses damage in DNA (pink) and triggers a protective response. The most commonly mutated gene in cancer, p53, works by shifting cell metabolism in a way that alters a cell’s identity, researchers at the Sloan Kettering Institute have discovered.
What is the main goal of genomics?
The aim of functional genomics is to describe and understand the pattern of gene expression. It has been established that gene expression is influenced by the expression of many genes.
What is genomic data used for?
Genomic data refers to the genome and DNA data of an organism. They are used in bioinformatics for collecting, storing and processing the genomes of living things. Genomic data generally require a large amount of storage and purpose-built software to analyze.
What does a genome test tell you?
A genomic test looks more widely at what's called your genome -- your full set of genetic "code" or instructions. It checks for gene mutations in your cancer to predict how it might act. A genomic test could help your doctor tell how fast-growing your cancer might be and how likely it is to spread.
What is the benefit of genome sequencing?
The primary purpose of sequencing one's genome is to obtain information of medical value for future care. Genomic sequencing can provide information on genetic variants that can lead to disease or can increase the risk of disease development, even in asymptomatic people.
What is genomics data analysis?
The Genomics Data Analysis XSeries is an advanced series that will enable students to analyze and interpret data generated by modern genomics technology. Using open-source software, including R and Bioconductor, you will acquire skills to analyze and interpret genomic data.
What is the difference between genomic and genetic?
The main difference between genomics and genetics is that genetics scrutinizes the functioning and composition of the single gene where as genomics addresses all genes and their inter relationships in order to identify their combined influence on the growth and development of the organism.
What is genomic sequence data?
Abstract. The Genome Sequence DataBase (GSDB) is a database of publicly available nucleotide sequences and their associated biological and bibliographic information.
How does genomics work?
Genomics is the study of whole genomes of organisms, and incorporates elements from genetics. Genomics uses a combination of recombinant DNA, DNA sequencing methods, and bioinformatics to sequence, assemble, and analyse the structure and function of genomes.
What is a genome?
An organism's complete set of DNA is called its genome. Virtually every single cell in the body contains a complete copy of the approximately 3 billion DNA base pairs, or letters, that make up the human genome.
Why is the Human Genome Project important?
The Human Genome Project was designed to generate a resource that could be used for a broad range of biomedical studies. One such use is to look for the genetic variations that increase risk of specific diseases, such as cancer, or to look for the type of genetic mutations frequently seen in cancerous cells. More research can then be done to fully understand how the genome functions and to discover the genetic basis for health and disease.
What is DNA sequencing?
Sequencing simply means determining the exact order of the bases in a strand of DNA. Because bases exist as pairs, and the identity of one of the bases in the pair determines the other member of the pair, researchers do not have to report both bases of the pair.
What is the Human Genome Project?
The Human Genome Project, which was led at the National Institutes of Health (NIH) by the National Human Genome Research Institute, produced a very high-quality version of the human genome sequence that is freely available in public databases. That international project was successfully completed in April 2003, under budget and more than two years ahead of schedule.
What are the implications for medical science?
These were conditions, such as sickle cell anemia, which have very simple, predictable inheritance patterns because each is caused by a change in a single gene.
What is DNA made of?
DNA molecules are made of two twisting, paired strands, often referred to as a double helix.
How do genes make proteins?
Located on 23 pairs of chromosomes packed into the nucleus of a human cell, genes direct the production of proteins with the assistance of enzymes and messenger molecules. Specifically, an enzyme copies the information in a gene's DNA into a molecule called messenger ribonucleic acid (mRNA). The mRNA travels out of the nucleus and into the cell's cytoplasm, where the mRNA is read by a tiny molecular machine called a ribosome, and the information is used to link together small molecules called amino acids in the right order to form a specific protein.
What is the study of the genome?
The genome includes both the genes (the coding regions) and the noncoding DNA, as well as mitochondrial DNA and chloroplast DNA. The study of the genome is called genomics .
When was the genome created?
The term genome was created in 1920 by Hans Winkler, professor of botany at the University of Hamburg, Germany. The Oxford Dictionary suggests the name is a blend of the words gene and chromosome. However, see omics for a more thorough discussion.
How much variation does the eukaryotic genome show?
The amount is even more than what is necessary for DNA protein-coding and noncoding genes due to the fact that eukaryotic genomes show as much as 64,000-fold variation in their sizes. However, this special characteristic is caused by the presence of repetitive DNA, and transposable elements (TEs).
Why are there so many different genome sizes?
The main reason why there is such a big variety of sizes is due to the presence of transposable elements. TEs are known to contribute to a significant change in a cell's mass of DNA.
What is the sequence of DNA?
A genome sequence is the complete list of the nucleotides (A, C, G, and T for DNA genomes) that make up all the chromosomes of an individual or a species. Within a species, the vast majority of nucleotides are identical between individuals, but sequencing multiple individuals is necessary to understand the genetic diversity.
How many pairs of chromosomes are there in an eukaryotic genome?
The number of chromosomes varies widely from Jack jumper ants and an asexual nemotode, which each have only one pair, to a fern species that has 720 pairs.
When was the first eukaryotic genome published?
A few months later, the first eukaryotic genome was completed, with sequences of the 16 chromosomes of budding yeast Saccharomyces cerevisiae published as the result of a European-led effort begun in the mid-1980s.
How does my genome work?
The same goes for your genome. The letters of your genome combine in different ways to spell out specific instructions.
How does genomics impact everyday life?
As technology advances and we learn more about how the genome works, information about our genomes is quickly becoming part of our everyday life. Emerging technologies give us the ability to read someone’s genome sequence. Having this information can lead to more questions about what genomics means for ourselves, our family members and society.
What is genetic testing?
Genetic testing consists of the processes and techniques used to determine details about your DNA. Depending on the test, it may reveal some information about your ancestry and the health of you and your family.
How many chromosomes are in a human?
Every fungus, plant, and animal has a set number of chromosomes. For example, humans have 46 chromosomes (23 pairs), rice plants have 24 chromosomes, and dogs have 78 chromosomes. Human Chromosomes.
What is DNA made of?
DNA is read like a code. This code is made up of four types of chemical building blocks, adenine, thymine, cytosine, and guanine, abbreviated with the letters A, T, C, ...
What is the operating manual of a cell?
Your genome is the operating manual containing all the instructions that helped you develop from a single cell into the person you are today. It guides your growth, helps your organs to do their jobs, and repairs itself when it becomes damaged. And it’s unique to you.
What is the name of the word for all your DNA?
Genome is a fancy word for all your DNA. From potatoes to puppies, all living organisms have their own genome. Each genome contains the information needed to build and maintain that organism throughout its life.
Why is it important to study the human genome?
Basic research exploring human and nonhuman genomes is critical to help scientists understand the basic biology underlying disease, as well as to discover new possible therapeutic targets.
Why is genome editing important?
Researchers rely on genome editing tools as a way to explore the connection between genotype (genes) and phenotype (traits). A typical study might be to model human disease in mice by deleting or editing certain genes that are thought to contribute to the disease. This approach can help researchers determine if specific changes made to the genome contribute to the disease. It can also lead to the creation of "disease models," or laboratory animals that mimic human diseases and can be studied to test new therapies.
What are the challenges of gene therapy?
One other challenge for gene therapy is that there is still much to learn about which genes are involved in most diseases and how different changes in these genes affect a person's risk of getting a disease. ...
Can germline therapy prevent inheritance?
These changes would then be passed down from generation to generation. Germline therapy could potentially prevent inheritance of diseases.
Can genome editing be used for cancer?
Many diseases from cancer to asthma have genetic bases. Through the application of genome editing technologies , physicians might eventually be able to prescribe targeted gene therapy to make corrections to patient genomes and prevent, stop, or reverse disease.
Can genome editing be used to treat diseases?
It can also lead to the creation of "disease models," or laboratory animals that mimic human diseases and can be studied to test new therapies. In the clinic, there are proposals to use genome editing as a treatment for disease. Many diseases from cancer to asthma have genetic bases.
Can genetics be used to treat cancer?
Through the application of genome editing technologies, physicians might eventually be able to prescribe targeted gene therapy to make corrections to patient genomes and prevent, stop, or reverse disease.
How Are Genomes Used?
Once scientists know the order of the DNA letters within a genome, they can use the map to figure out where they need to go to start their research.
What is the blue print of DNA?
DNA: deoxyribonucleic acid is the information "blue-print" of the cell. It is a nucleic acid and is made from building blocks called nucleotides. This genetic information is passed from parent to child... more (link is external)
What is the study of the structure and function of the genome called?
The study of the structure and function of the genome is called genomics. DNA is a polymer - a large and complex molecule. The monomers that make it are called nucleotides. DNA is made up of two strands forming a twisted ladder structure called a double helix. The nucleotides are arranged along the DNA to form a code.
What is a gene?
A gene is a small section of genetic code in the DNA of a chromosome. Each gene codes for a particular sequence of amino acids in order to make a specific protein. It is a unit of heredity, and may be copied and passed on to the next generation.
Where is the genetic material of eukaryotes?
Genetic material for eukaryotes is in their nucleus. Prokaryotes such as bacteria do not have nuclei. Their genetic material is in the cytoplasm and includes small loops of DNA called plasmids.
Does DNA code for proteins?
Most DNA - 98.5 per cent in humans - does not code for proteins. Instead, it seems to be important in controlling gene expression. This means it controls when genes are 'read' to make proteins. The genes and non-coding DNA are mixed together in the chromosomes.
Why do scientists use genome editing?
One way that scientists use genome editing is to investigate different diseases that affect humans.
How does genome editing work?
Genome editing technologies enable scientists to make changes to DNA, leading to changes in physical traits, like eye color, and disease risk. Scientists use different technologies to do this. These technologies act like scissors, cutting the DNA at a specific spot. Then scientists can remove, add, or replace the DNA where it was cut.
Why do scientists edit the genome?
They edit the genomes of animals, like mice and zebrafish, because animals have many of the same genes as humans. For example, mice and humans share about 85 percent of their genes! By changing a single gene or multiple genes in a mouse, scientists can observe how these changes affect the mouse's health and predict how similar changes in human genomes might affect human health.
What is genome editing?
Genome editing is a method that lets scientists change the DNA of many organisms, including plants, bacteria, and animals. Editing DNA can lead to changes in physical traits, like eye color, and disease risk. Scientists use different technologies to do this.
How much of the human genome does a mouse share?
For example, mice and humans share about 85 percent of their genes! By changing a single gene or multiple genes in a mouse, scientists can observe how these changes affect the mouse's health and predict how similar changes in human genomes might affect human health.
When was genome editing invented?
The first genome editing technologies were developed in the late 1900s. More recently, a new genome editing tool called CRISPR, invented in 2009, has made it easier than ever to edit DNA. CRISPR is simpler, faster, cheaper, and more accurate than older genome editing methods. Many scientists who perform genome editing now use CRISPR. Overview. ...
When was the first genome editing technology developed?
Then scientists can remove, add, or replace the DNA where it was cut. The first genome editing technologies were developed in the late 1900s.
What are the applications of genome sequencing?
Here are some of the applications of genome sequencing. 1. Diagnostics and Medicine. DNA sequencing has elaborate applications in screening the risk of genetic diseases, gene therapy-based treatments, genetic engineering, and gene manipulation.
What is genome sequencing?
Updated March 20, 2019. Genome sequencing refers to the process of determining the order of the nucleotides bases— adenine, guanine, cytosine, and thymine in a molecule of DNA or the genome of an organism. The methods of sequencing have become a game-changer in modern ...
How does Illumina sequencing work?
Illumina sequencing technology uses the principle of sequencing by synthesis. Sequencing templates are immobilized on a proprietary flow cell surface. Unlabeled nucleotides and enzymes are added to initiate solid-phase bridge amplification. The enzyme incorporates nucleotides to build double-stranded bridges on the solid-phase substrate. Denaturation leaves single-stranded templates anchored to the substrate. Several million dense clusters of double-stranded DNA are generated in each channel of the flow cell. The first sequencing cycle begins by adding four labeled reversible terminators, primers, and DNA polymerase. After laser excitation, the emitted fluorescence from each cluster is captured, and the first base is identified. The next cycle repeats the incorporation of four labeled reversible terminators, primers, and DNA polymerase. After laser excitation, the image is captured as before, and the identity of the second base is recorded. The sequencing cycles are repeated to determine the sequence of bases in a fragment, one base at a time. The data are aligned and compared to a reference, and sequencing differences are identified.
What is the purpose of a dNTP label?
During each sequencing cycle, a single labeled dNTP is added to the nucleic acid chain. The nucleotide label serves as a terminator for polymerization, so after each dNTP incorporation, the fluorescent dye is imaged to identify the base and then enzymatically cleaved to allow incorporation of the next nucleotide.
What is the first generation sequencing method?
The first generation sequencing methods were the earliest sequencing technologies developed and are known as the basic methods of sequencing. There were particularly two significant sequencing techniques in the first generation. 1. Maxam-Gilbert Sequencing.
Why is sequencing important in agriculture?
Sequencing of microorganisms to engineer resistant genes in crops. Mapping and whole-genome sequencing of food plants to increase productivity and nutritional contents as well as environmental tolerance .
When was DNA sequencing first used?
Source. The earliest successful attempt of DNA sequencing dates back to the early 1970s.
What is the purpose of the completed sequence of the human genome?
Effectively, the completed sequence of the human genome provides an instruction manual for making and maintaining the human body. The task for scientists now is to read the contents of this instruction manual and work out how all the parts fit together and what it all means. The completion of the human genome is just the starting point for a century or more of new discoveries. A number of genome sequencing projects have been set up since the Human Genome Project to start to understand genome biology. You can read about some of them below.
How many genomes were studied in 2010?
In 2010, the Wellcome Trust launched a study of 10,000 human genomes in the UK, which aimed to analyse the DNA of one in every 6,000 individuals in the UK. Scientists expected the UK10K project to uncover many rare genetic variants that are important in human disease, giving a much deeper picture of genetics to help other studies, both in the UK and around the world.
What was the first study to sequence and compare the genomes of large numbers of people?
Studies like the 1000 Genomes Project and UK10K were the first studies to sequence and compare the genomes of large numbers of people.
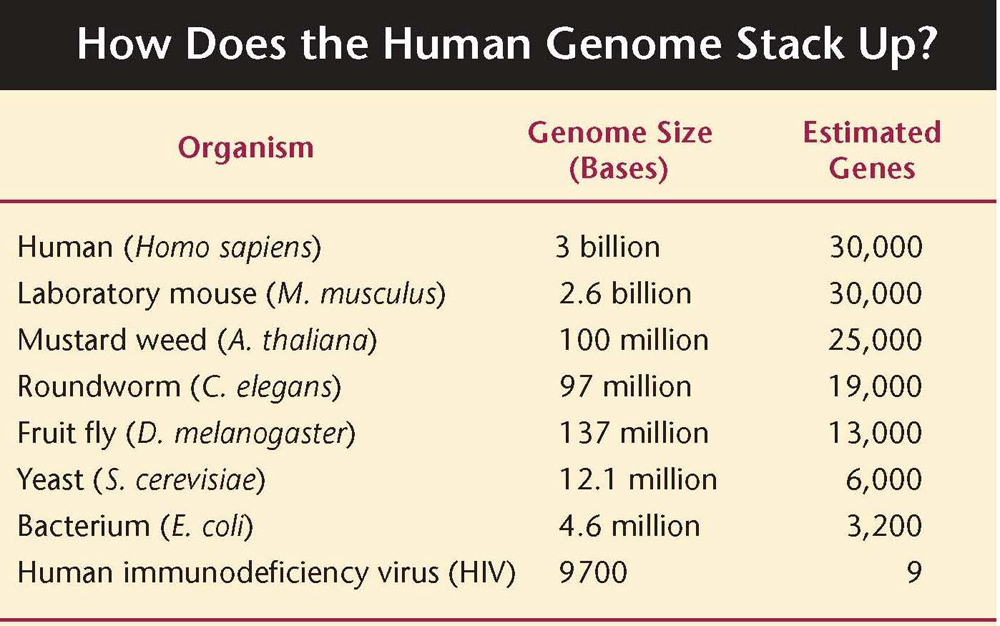
How many genes are in the human genome?
As well as confirming the number of genes that carry the codes to make proteins (around 21,000), they identified another 9,000 genes that have important effects in our cells. They also announced that 80 per cent of the human genome sequence is close to regions that control biological function; this meant that most of what was once considered “junk DNA”, the non-coding DNA in the human genome, is actually functional.
What is the pattern of blocks on chromosomes called?
The pattern of these blocks in our chromosomes is called a haplotype. A HapMap is a map of where these haplotype blocks are located on the chromosome. This is useful because, by comparing the HapMap of different individuals, scientists can then identify key areas of genetic variation.
Why is it important to have a catalogue of genes?
Having a catalogue like this has made it much easier to establish connections between particular genes and disease.
When was the 1000 genomes project started?
The 1000 Genomes Project, which launched in 2008, was the first project to aim to sequence the genomes of a large number of people (at least 1,000), to provide a comprehensive resource on human genetic variation.

Overview
Viral genomes
Viral genomes can be composed of either RNA or DNA. The genomes of RNA viruses can be either single-stranded RNA or double-stranded RNA, and may contain one or more separate RNA molecules (segments: monopartit or multipartit genome). DNA viruses can have either single-stranded or double-stranded genomes. Most DNA virus genomes are composed of a single, linear molecule of DNA, but some are made up of a circular DNA molecule.
Origin of term
The term genome was created in 1920 by Hans Winkler, professor of botany at the University of Hamburg, Germany. The Oxford Dictionary suggests the name is a blend of the words gene and chromosome. However, see omics for a more thorough discussion. A few related -ome words already existed, such as biome and rhizome, forming a vocabulary into which genome fits systematically.
Defining the genome
It's very difficult to come up with a precise definition of "genome." It usually refers to the DNA (or sometimes RNA) molecules that carry the genetic information in an organism but sometimes it is difficult to decide which molecules to include in the definition; for example, bacteria usually have one or two large DNA molecules (chromosomes) that contain all of the essential genetic material but they also contain smaller extrachromosomal plasmid molecules that carry important genetic …
Sequencing and mapping
A genome sequence is the complete list of the nucleotides (A, C, G, and T for DNA genomes) that make up all the chromosomes of an individual or a species. Within a species, the vast majority of nucleotides are identical between individuals, but sequencing multiple individuals is necessary to understand the genetic diversity.
In 1976, Walter Fiers at the University of Ghent (Belgium) was the first to establish the complete n…
Prokaryotic genomes
Prokaryotes and eukaryotes have DNA genomes. Archaea and most bacteria have a single circular chromosome, however, some bacterial species have linear or multiple chromosomes. If the DNA is replicated faster than the bacterial cells divide, multiple copies of the chromosome can be present in a single cell, and if the cells divide faster than the DNA can be replicated, multiple replication of the chromosome is initiated before the division occurs, allowing daughter cells to i…
Eukaryotic genomes
Eukaryotic genomes are composed of one or more linear DNA chromosomes. The number of chromosomes varies widely from Jack jumper ants and an asexual nemotode, which each have only one pair, to a fern species that has 720 pairs. It is surprising the amount of DNA that eukaryotic genomes contain compared to other genomes. The amount is even more than what is necessary for DNA protein-coding and noncoding genes due to the fact that eukaryotic genome…
Genome size
Genome size is the total number of the DNA base pairs in one copy of a haploid genome. Genome size varies widely across species. Invertebrates have small genomes, this is also correlated to a small number of transposable elements. Fish and Amphibians have intermediate-size genomes, and birds have relatively small genomes but it has been suggested that birds lost a substantial portion of their genomes during the phase of transition to flight. Before this loss, DNA methylati…