
How do you Digest a plasmid?
Procedure Select restriction enzymes to digest your plasmid. Determine an appropriate reaction buffer by reading the instructions for your enzyme. In a 1.5mL tube combine the following: DNA Mix gently by pipetting. Incubate tube at appropriate temperature (usually 37 °C) for 1 hour.
How much plasmid backbone do I need for digestion?
We recommend 1.5-2μg of insert and 1μg of plasmid backbone. It is also critical that as much of the backbone plasmid as possible be cut with both enzymes, and therefore it is important that the digest go until completion. The time required for complete digestion varies for different enzymes.
How to test individual plasmid clones before using DNA sequencing?
This is frequently done after performing either PCR - or restriction enzyme -based cloning to test individual clones before use of more expensive forms of plasmid verification, such as DNA sequencing. In the example above, digestion with enzyme RE1 will linearize the 6200bp plasmid into one single 6200bp fragment.
How do I screen and verify my plasmids?
This method is time-intensive, so we recommend a variety of ways to screen and verify your plasmids. Here, we'll cover restriction digest analysis. Diagnostic digests can be used to confirm the rough structure of the plasmid based on the predicted sizes and organization of different features within the plasmid.
What does digest a plasmid mean?
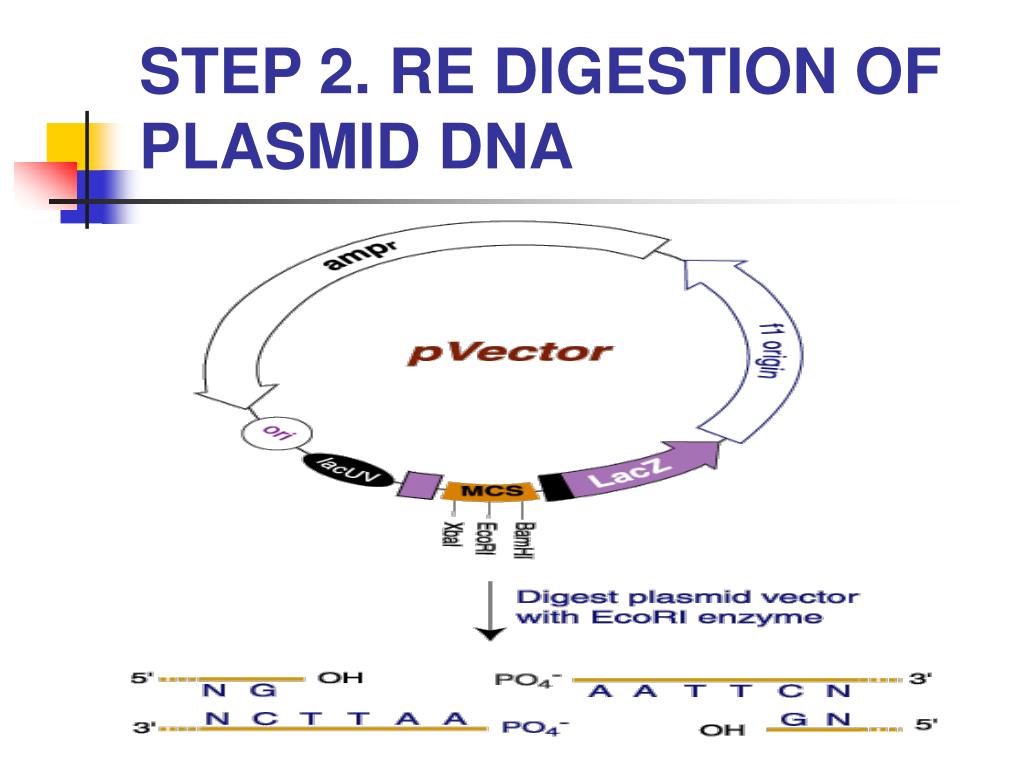
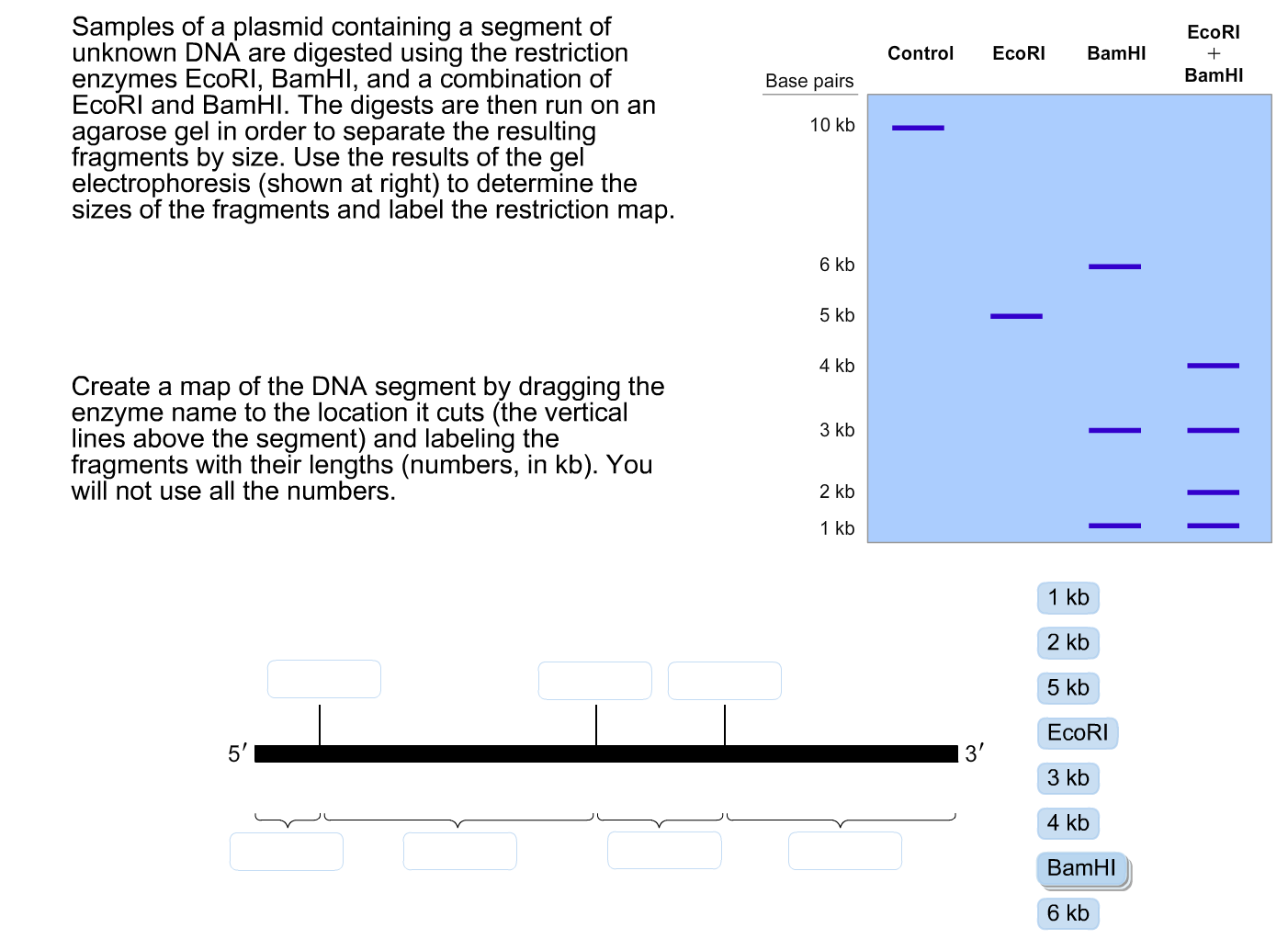
The goal of a diagnostic digest is to cut your plasmid into specific sized pieces and analyze the resulting fragments by gel electrophoresis. The pattern of the fragments on the gel can indicate if the plasmid contains the expected size insert.
How do you digest plasmid?
ProcedureSelect restriction enzymes to digest your plasmid. ... Determine an appropriate reaction buffer by reading the instructions for your enzyme. ... In a 1.5mL tube combine the following: ... Mix gently by pipetting.Incubate tube at appropriate temperature (usually 37 °C) for 1 hour.More items...
What does Digest mean in genetics?
To cut DNA molecules with one or more Restriction endonucleases. Tags: Molecular Biology.
What is the purpose of DNA digestion?
Restriction digestion is usually used to prepare a DNA fragment for subsequence molecular cloning, as the procedure allows fragments of DNA to be pieced together like building blocks via ligation.
What is the difference between digested and undigested DNA?
Completely digested plasmid DNA usually show only a single band, a linear form of the plasmid, in its lane with the expected size. Undigested plasmid may have two forms show up in its lane: CCC dimer and CCC monomer forms.
What is the steps in restriction digestion?
Protocol for DNA Digestion with a Single Restriction Enzyme Incubate the reaction at digestion temperature (usually 37 °C) for 1 hour. Stop the digestion by heat inactivation (65 °C for 15 minutes) or addition of 10 mM final concentration EDTA. The digested DNA is ready for use in research applications.
Do we digest DNA?
Basically, DNA, like proteins and complex carbohydrates, gets broken down into pieces – this is what digestion is all about. Your teeth mash it up and enzymes throughout your digestive tract cut it to pieces.
What is meant by digestion in biology?
Digestion is defined as the process of breaking down large, insoluble molecules of food into smaller, water-soluble molecules which can then be readily absorbed by the body. Digestion is one among many life processes observed in nearly all living organisms.
What is digestion and absorption?
Digestion and absorption occur in the digestive tract. After the nutrients are absorbed, they are available to all cells in the body and are utilized by the body cells in metabolism. The digestive system prepares nutrients for utilization by body cells through six activities, or functions.
Why must DNA be digested with restriction enzymes before electrophoresis?
Explanation: There exist an enzyme, called restriction enzyme, that can identify a particular nucleotide sequence, called restriction sites, and perform cleaving operation. This process separates genetic material into smaller fragments which may contain gene(s) of interest.
Can digested DNA be stored?
The product of restriction digestion can be easily stored at -20 C. At 4 C it would be fine but to ensure that there is no activity and no star activity it is recommended to keep it at -20 C.
Why partial digestion of DNA is important?
Additionally, molecular biologists use partial restriction enzyme reaction to fragment or “digest” genomic DNA for Southern blot analysis. By carrying out partial restriction enzyme digests with the genomic DNA, they can generate larger fragment sizes.
How does undigested plasmid run on gel?
Undigested plasmid may have two forms show up in its lane: CCC dimer and CCC monomer forms. The dimer forms, due to their larger and doubling size compared to monomers, usually move slower than the monomers. Therefore, it will appear higher in a gel than a monomer.
What enzyme digests DNA?
Restriction EndonucleasesRestriction Digestion is the process of cutting DNA molecules into smaller pieces with special enzymes called Restriction Endonucleases (sometimes just called Restriction Enzymes or RE's).
Can you digest PCR product directly?
Frequently, a PCR product must be further manipulated by cleavage with restriction enzymes. For convenience, restriction enzyme digestion can be performed directly in the PCR mix without any purification of the DNA.
Can you store digested plasmid?
The product of restriction digestion can be easily stored at -20 C. At 4 C it would be fine but to ensure that there is no activity and no star activity it is recommended to keep it at -20 C.
How long to digest DNA for cloning?
For diagnostic digests, 1-2 hours is often sufficient. For digests with >1 µg of DNA used for cloning, it is recommended that you digest for at least 4 hours. *Pro-Tip* If you will be using the digested DNA for another application (such as a digestion with another enzyme in a different buffer), but will not be gel purifying it, ...
What is restriction enzyme digestion?
Restriction enzyme digestion takes advantage of naturally occurring enzymes that cleave DNA at specific sequences. There are hundreds of different restriction enzymes, allowing scientists to target a wide variety of recognition sequences. For a list of many commonly used restriction enzymes, visit NEB .
Why do enzymes need to be placed in an ice bucket immediately after removal from the freezer?
Restriction enzymes MUST be placed in an ice bucket immediately after removal from the -20 °C freezer because heat can cause the enzymes to denature and lose their function.
Why do enzymes cut sequences that are similar but not identical to their recognition sites?
Sometimes enzymes cut sequences which are similar, but not identical, to their recognition sites. This is due to "Star Activity" and can happen for a variety of reasons, including high glycerol concentration. Learn more at NEB's website about star activity.
How long to incubate DNA?
*Pro-Tip* Depending on the application and the amount of DNA in the reaction, incubation time can range from 45 mins to overnight. For diagnostic digests, 1-2 hours is often sufficient.
Can a plasmid be resistant to cleavage?
Plasmids grown in Dam or Dcm methylase positive strains will be resistant to cleavage at certain restriction sites. See NEB's table of methylation sensitive restriction sites. Sometimes enzymes cut sequences which are similar, but not identical, to their recognition sites.
Can you mix DNA with plasmids?
If you are digesting a large number of plasmids with the same enzyme (s) (for instance, in a diagnostic digest), you can create a "Master Mix" consisting of all of the reaction components except for the DNA. Aliquot your DNA into individual tubes and then add the appropriate amount of Master Mix to each tube. This will save you time and ensure consistency across the reactions.
What enzymes are needed for a diagnostic digest?
Before beginning your diagnostic digest, you will need to select a restriction enzyme or enzymes that cut your plasmid. Many DNA analysis tools, including Addgene’s Sequence Analyzer, allow you to identify which restriction sites are present in a given sequence. For a list of the commonly used commercially available restriction enzymes, see New England Biolabs' website .
What is diagnostic digest?
The goal of a diagnostic digest is to cut your plasmid into specific sized pieces and analyze the resulting fragments by gel electrophoresis. The pattern of the fragments on the gel can indicate if the plasmid contains the expected size insert.
What is restriction digest?
Given the variety of these enzymes and the unique sites they recognize, restriction digests have become the most widely used method scientists employ to selectively move a specific piece of DNA from one plasmid to another.
What is the enzyme that binds a plasmid to a new fragment?
An enzyme, DNA ligase, then covalently binds the plasmid to the new fragment thereby generating a complete, circular plasmid that can be easily maintained in a variety of biological systems. Read on for an in-depth breakdown of how to do perform restriction digests. Before beginning the restriction digest and ligation process, ...
What enzymes are used to cut a plasmid?
When cloning by restriction digest and ligation, you use restriction enzymes to cut open a plasmid (backbone) and insert a linear fragment of DNA (insert) that has been cut by compatible restriction enzymes. An enzyme, DNA ligase, then covalently binds the plasmid to the new fragment thereby generating a complete, circular plasmid that can be easily maintained in a variety of biological systems. Read on for an in-depth breakdown of how to do perform restriction digests.
How much DNA should be used in a ligation reaction?
We recommend around 100ng of total DNA in a standard ligation reaction. You ideally want a “recipient plasmid : insert ratio” of approximately 1:3. Since the number of base pairs for each varies, it is difficult to calculate this based on DNA concentration alone. One method is to conduct 2 ligations for each plasmid you are trying to create, with varying ratios of recipient plasmid to insert.
What enzymes are used to prevent re-circularization of the backbone plasmid?
If you are going to use only one restriction enzyme, or enzymes that have compatible overhangs or no overhangs after digestion, you will need to use a phosphatase to prevent re-circularization of the backbone plasmid (see below). You should treat your digested backbone plasmid with a phosphatase prior to the ligation step or prior to the gel purification step, depending on the phosphatase you choose. CIP (calf alkaline phosphatase ) or SAP (shrimp alkaline phosphatase ) are commonly used. Follow the manufacturer’s instructions.
How to make a circular plasmid?
In the ligation step, you mix your purified, cut backbone and insert in a single tube allowing the compatible overhangs generated by restriction digestion to anneal to one another and form a complete, circular plasmid. You then add DNA ligase to covalently link the fragments together at the expense of ATP (see below, covalent bonds are indicated in red).
What is the best way to treat digested backbone plasmid?
You should treat your digested backbone plasmid with a phosphatase prior to the ligation step or prior to the gel purification step, depending on the phosphatase you choose. CIP (calf alkaline phosphatase) or SAP (shrimp alkaline phosphatase) are commonly used. Follow the manufacturer’s instructions. 2.
What enzyme is used to make bonds between the insert and backbone?
Both the plasmid (blue, backbone) and the DNA sequence of interest (green, insert) are cut with restriction enzymes to generate compatible overhangs that allow them to bind. Ligase is used to make bonds between the insert and backbone covalent.
How long does it take for plasmids to digest?
It is also critical that as much of the recipient plasmid as possible be cut with both enzymes, and therefore it is important that the digest go at least 4 hours and as long as overnight. If you are going to use only one restriction enzyme, or enzymes that have compatible overhangs or no overhangs after digestion, ...
How to run a diagnostic digest?
After purifying the DNA, conduct a diagnostic restriction digest of 100-300ng of your purified DNA with the enzymes you used for the cloning. Run your digest on an agarose gel. You should see two bands, one the size of your vector and one the size of your new insert. If you used only one enzyme or used enzymes with compatible overhangs you will need to verify the orientation of your insert, so you may want to design a diagnostic digest for this purpose.
What is subcloning by restriction digest?
Subcloning by restriction digest is a commonly used lab technique. For the purposes of this tutorial we will discuss how to move a cDNA from one plasmid to another. However, the same technique can be used to move promoters, selectable markers, or any other DNA element between plasmids.
What is the purpose of DNA ligation?
Conduct a DNA Ligation to fuse your insert to your recipient plasmid.
How much DNA can you use for cloning?
For most standard cloning, you can transform 1-2μl of your ligation reaction into competent cells such as DH5alpha or TOP10. If using much less total DNA (<1ng) or if you are having trouble getting colonies, you might want to use higher competency cells. Additionally, if your final product is going to be very large (>10kb) you might want to use electro-competent cells instead of the more common chemically-competent cells.
Can you use PCR to flank oligos?
You have other options, such as: Adding desired restriction sites to flank your insert : You can use PCR Based Cloning and add restriction sites to the ends of your oligos. This will allow you to produce a version of your insert flanked by restriction sites compatible with the recipient plasmid's MCS.
Can DNA be moved from one vector to another?
The following technique can be used to easily move any piece of DNA from one vector to another as long as it is already bounded by restriction sites that are also present in the same orientation on your target vector. If you are not sure what vector to use, you can check out our Empty Backbone Reference .
How long does it take for a fast digest enzyme to digest?
We use FastDigest Enzymes in our lab. Guess what the Fast in the name stands for: they are really fast! We use about 100 ng of plasmid and digest them for 10 min. 37°C which is enough time to digest that amount of plasmid. Actually the protocol says that the fidelity of the enzyme is so high that it digests 1µg of phage DNA in 5 min.
What is a fast digest?
FastDigest is the Fermentas equivalent of NEB's HF enzymes that are all tweaked to work in a single buffer and have reduced star activity. I would bet a lot of money that NEB Buffer 4 and the Fermentas FastDigest buffer are interchangable, and since your NEB NheI works in NEB Buffer 4 I'd suggest performing a double digest in this (or in the FastDigest buffer, or both).
How many bands of plasmid should be on gel?
Usually, a uncut plasmid should show at least three bands on gel. The top band most likely is the nicked/linearized plasmid. A mid band is relatively thicker which is the supercoiled plasmid. And if there is a lower band most likely the single circular formation of the plasmid.
How many ug of enzymes to digest excised fragment?
Alternatively, if your goal is to clone the excised fragment I'd recommend starting wtih 5-10 ug plasmid and digest with 20-60U of each enzyme (but would still digest for the same length of time).
How good is an enzyme cut?
Finally, how good an enzyme cuts is not only a matter of time. It's also a matter of how much DNA you have, how much enzyme, and how clean the reaction is. Good luck.
Why join Researchgate?
Join ResearchGate to find the people and research you need to help your work.
Do non-fast digest enzymes work in FD buffer?
Concerning the double digestion: we made the observation that even non-Fastdigest enzymes from other manufacturers work usually very well in the FD-buffer, so that we always try the FD buffer before anything else when making double digests.
How many base pairs are in a plasmid?
As you see from the map center, the size of the linearized plasmid is 4361 base pairs. Before you start working with any plasmid, it is advisable to linearize it by cutting with a unique restriction enzyme to check that the advertised size is roughly the same as expected.
What are the transcription promoters and terminators of plasmids?
In addition to genes, plasmids often include transcription promoters and terminators derived from E.coli phages. Promoters from phages SP6 and T7 are often used for in vitro RNA amplification. They require phage polymerases and are therefore inactive in vivo.
What are the two antibiotic resistance genes in pBR322?
pBR322 has two antibiotic resistance genes: tet (tetracycline resistance) and amp (ampicillin resistance). These genes encode an efflux pump ( tetR) and beta-lactamase ( ampR) to excrete tetracycline and ampicillin from the cell, respectively. Tet and amp are read in different directions.
Can you have two plasmids with the same oi?
Actually, you can maintain two plasmids with the same ori in one bacteria as long as they have different antibiotic resistance genes and you have both antibiotics in the media.
Can a plasmid replicate?
Once a plasmid is unable to replicate, it is useless. The other thing to remember about ori is that plasmids with the same origin are often incompatible. This means that you will not be able to maintain two pBR323- derived vectors in one cell even if they have genes for different antibiotic resistances on them.
Why is my enzyme not digesting?
If your enzyme is active and digests the control DNA and the reaction is set up using optimal conditions, but you still see issues with digestion, it might be because the enzyme is inhibited by methylation of the template DNA.
Why is it so difficult to cleave a double digest?
If you are trying to perform a double digest with two enzymes in the multiple cloning site, efficient cleavage may be difficult if the two recognition sites are too close together. One enzyme may physically block access of the second enzyme to its respective site. Inefficient cleavage is also related to the previously described proximity of the recognition site to DNA ends. After one enzyme cuts, there may not be enough bases flanking the second site for the second enzyme to bind and effect cleavage.
Why are PCR fragments incomplete?
PCR fragments: Incomplete or no digestion of PCR products may be due to the proximity of the recognition site to the end of the DNA fragment. Some restriction enzymes require additional flanking bases for efficient DNA binding and cleavage ( Figure 4 ). Because recognition sites are often introduced at the ends of PCR fragments and/or primers, ...
How many bases does a paei need to cleave DNA?
For example, PasI can cleave DNA even if the recognition site is at the very end of the fragment, while PaeI requires at least 5 additional bases for optimal digestion ( Figure 5 ).
What are the issues with DNA methylation?
Issue 1. Incomplete or no digestion due to inactive enzyme. Issue 2. Incomplete or no digestion due to suboptimal reaction conditions. Issue 3. Incomplete or no digestion due to enzyme activity blocked by DNA methylation. Issue 4. Incomplete or no digestion due to the structure of substrate DNA. Issue 5.
How to test enzyme activity?
Test the enzyme for activity by setting up a control reaction with 1 µg of standard control DNA (e.g., lambda DNA), where you know that the DNA quality is high and the expected banding pattern ( Figure 1 ).
What can inhibit enzymes?
Double check the optimal reaction temperature for the enzymes being used, and control for evaporation during incubation as an increased salt concentration in the buffer can inhibit enzyme performance.
Example
Treatment
- If you are going to use only one restriction enzyme, or enzymes that have compatible overhangs or no overhangs after digestion, you will need to use a phosphatase to prevent re-circularization of the backbone plasmid (see below). You should treat your digested backbone plasmid with a phosphatase prior to the ligation step or prior to the gel purification step, depending on the phos…
Resources
- For more information on these stains see the Bitesize Bio Blog and their associated manufacturers websites. Additional Resources: Additional Resources on the Addgene Blog: Resources on Addgene.org
Risks
- These stains require you to either stain your gel after you run your samples or add the stain as the gel is being made (post or pre run in the table above, respectively). Some of the above stains require you to visualize your DNA using UV light please note that UV light can damage DNA and that proper personal protective equipment should be worn when visualizing using UV as it can c…
Preparation
- Once you have cut out and purified your insert and recipient plasmid backbone bands away from the gel via your favorite gel purification method, it is important to determine the concentration of recovered DNA as this will be useful for the ligation step.
Use
- In the ligation step, you mix your purified, cut backbone and insert in a single tube allowing the compatible overhangs generated by restriction digestion to anneal to one another and form a complete, circular plasmid. You then add DNA ligase to covalently link the fragments together at the expense of ATP (see below, covalent bonds are indicated in...
Prevention
- Transform your ligation reaction into your bacterial strain of choice. Follow the manufacturers instructions for your competent cells.
Results
- Sample results indicative of successful and unsuccessful ligations are indicated below. A successful ligation will have few colonies on the backbone alone plate and many colonies on the backbone + insert plate (or at least more colonies than the backbone alone plate). Unsuccessful ligations will usually result in few colonies on both plates (unsuccessful 1), in a vector alone plat…