What is the abbreviation for malate dehydrogenase?
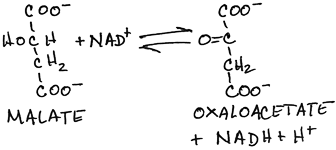
For other uses, see Malate dehydrogenase (disambiguation). Malate dehydrogenase ( EC 1.1.1.37) ( MDH) is an enzyme that reversibly catalyzes the oxidation of malate to oxaloacetate using the reduction of NAD + to NADH. This reaction is part of many metabolic pathways, including the citric acid cycle.
What is the role of malate dehydrogenase in the citric acid cycle?
In the citric acid cycle, malate dehydrogenase is responsible for catalyzing the regeneration of oxaloacetate This reaction occurs through the oxidation of hydroxyl group on malate and reduction of NAD +.
What is malate dehydrogenase in the liver?
Malate Dehydrogenase. Malate dehydrogenase (MDH) is involved in the Krebs cycle, catalyzing the reversible transformation of malate into oxaloacetate. It is also a leakage marker released into serum after tissue damage. It is highly abundant in liver followed by heart, skeletal muscle, and brain.
What is malate dehydrogenase (NADP +)?
This reaction is part of many metabolic pathways, including the citric acid cycle. Other malate dehydrogenases, which have other EC numbers and catalyze other reactions oxidizing malate, have qualified names like malate dehydrogenase (NADP +) . Several isozymes of malate dehydrogenase exist. There are two main isoforms in eukaryotic cells.
See more

Where is malate dehydrogenase used?
liverMalate dehydrogenase (MDH) is involved in the Krebs cycle, catalyzing the reversible transformation of malate into oxaloacetate. It is also a leakage marker released into serum after tissue damage. It is highly abundant in liver followed by heart, skeletal muscle, and brain.
What is the role of malate dehydrogenase in gluconeogenesis?
Malate dehydrogenase is also involved in gluconeogenesis, the synthesis of glucose from smaller molecules. Pyruvate in the mitochondria is acted upon by pyruvate carboxylase to form oxaloacetate, a citric acid cycle intermediate.
Why is MDH important?
Malate dehydrogenase (MDH), a key enzyme in the tricarboxylic acid cycle (TCA), plays important metabolic roles in aerobic energy producing pathways and in malate shuttle (Figure 1) [5]. The enzyme is widely distributed in animals, higher plants, and microorganisms.
What type of reaction does malate dehydrogenase catalyze?
Malate dehydrogenases catalyzes the interconversion of malate to oxaloacetate. In the citric acid cycle, malate dehydrogenase is responsible for catalyzing the regeneration of oxaloacetate This reaction occurs through the oxidation of hydroxyl group on malate and reduction of NAD+.
What type of enzyme is malate dehydrogenase?
Malate dehydrogenase (MDH) is a predominately periportal enzyme that is expressed highly in the extra-mitochondrial cytoplasm of the liver, although 10% of MDH has been reported in the mitochondria [23]. It is an enzyme in the citric acid cycle that catalyzes the reversible conversion of malate into oxaloacetate.
What is the reaction that MDH catalyzes?
Malate dehydrogenase (MDH, EC: 1.1. 1.37) reversibly catalyzes the oxidation of L-malate (MAL) to oxaloacetate (OAA), reducing NAD to NADH in the process. In eukaryotes this enzyme is expressed as mitochondrial (mMDH) and the cytosolic (cMDH) isoforms.
Why is malate dehydrogenase unfavorable?
The malate dehydrogenases are catalyzing an energetically unfavorable (non-spontaneous) reaction as the ΔG0 is larger than 0 7-9. MDH1 and MDH2 are encoded by distinct genes and differ in their specificity for the NAD+/NADH. Both MDH isoenzymes exist in physiological conditions as dimers with identical subunits 8, 10.
How is malate converted to pyruvate?
In the malate-pyruvate pathway, malate is exported from the mitochondria to the cytosol and converted directly to pyruvate by ME1.
What is gluconeogenesis pathway?
Gluconeogenesis is the pathway by which glucose is synthesized from non-carbohydrate metabolites. The principal gluconeogenic precursors are pyruvate and lactate, certain gluconeogenic amino acids, and glycerol, which is derived mainly from fat metabolism.
What is the reaction that MDH catalyzes?
Malate dehydrogenase (MDH, EC: 1.1. 1.37) reversibly catalyzes the oxidation of L-malate (MAL) to oxaloacetate (OAA), reducing NAD to NADH in the process. In eukaryotes this enzyme is expressed as mitochondrial (mMDH) and the cytosolic (cMDH) isoforms.
How is malate converted to pyruvate?
In the malate-pyruvate pathway, malate is exported from the mitochondria to the cytosol and converted directly to pyruvate by ME1.
Why is malate dehydrogenase unfavorable?
The malate dehydrogenases are catalyzing an energetically unfavorable (non-spontaneous) reaction as the ΔG0 is larger than 0 7-9. MDH1 and MDH2 are encoded by distinct genes and differ in their specificity for the NAD+/NADH. Both MDH isoenzymes exist in physiological conditions as dimers with identical subunits 8, 10.
Where is malate dehydrogenase found?
Several isozymes of malate dehydrogenase exist. There are two main isoforms in eukaryotic cells. One is found in the mitochondrial matrix, participating as a key enzyme in the citric acid cycle that catalyzes the oxidation of malate.
What is the active site of malate dehydrogenase?
The active site of malate dehydrogenase is a hydrophobic cavity within the protein complex that has specific binding sites for the substrate and its coenzyme, NAD +. In its active state, MDH undergoes a conformational change that encloses the substrate to minimize solvent exposure and to position key residues in closer proximity to the substrate. The three residues in particular that comprise a catalytic triad are histidine (His-195), aspartate (Asp-168), both of which work together as a proton transfer system, and arginines (Arg-102, Arg-109, Arg-171), which secure the substrate.
What happens when histidine is protonated?
Specifically, when the histidine is protonated, the His residue can form a hydrogen bond with the substrate's carbonyl oxygen, which shifts electron density away from the oxygen and makes it more susceptible to nucleophilic attack by hydride. This promotes the binding of malate dehydrogenase to these substrates.
What is the role of malate dehydrogenase in gluconeogenesis?
Malate dehydrogenase is also involved in gluconeogenesis, the synthesis of glucose from smaller molecules. Pyruvate in the mitochondria is acted upon by pyruvate carboxylase to form oxaloacetate, a citric acid cycle intermediate. In order to get the oxaloacetate out of the mitochondria, malate dehydrogenase reduces it to malate, and it then traverses the inner mitochondrial membrane. Once in the cytosol, the malate is oxidized back to oxaloacetate by cytosolic malate dehydrogenase. Finally, phosphoenolpyruvate carboxykinase (PEPCK) converts oxaloacetate to phosphoenolpyruvate (PEP).
What is the role of malate dehydrogenase in the citric acid cycle?
In the citric acid cycle, malate dehydrogenase is responsible for catalyzing the regeneration of oxaloacetate This reaction occurs through the oxidation of hydroxyl group on malate and reduction of NAD +. The mechanism of the transfer of the hydride ion ...
How does malate dehydrogenase get oxaloacetate out of the mitochondria?
In order to get the oxaloacetate out of the mitochondria, malate dehydrogenase reduces it to malate, and it then traverses the inner mitochondrial membrane. Once in the cytosol, the malate is oxidized back to oxaloacetate by cytosolic malate dehydrogenase.
What enzyme is responsible for oxidation of malate to oxaloacetate?
proteins. Malate dehydrogenase ( EC 1.1.1.37) ( MDH) is an enzyme that reversibly catalyzes the oxidation of malate to oxaloacetate using the reduction of NAD + to NADH. This reaction is part of many metabolic pathways, including the citric acid cycle. Other malate dehydrogen ases, which have other EC numbers and catalyze other reactions oxidizing ...
What is the function of malate dehydrogenase?
Malate dehydrogenase (MDH) (EC 1.1.1.37) catalyzes the conversion of oxaloacetate and malate. This reaction is important in cellular metabolism, and it is coupled with easily detectable cofactor oxidation/reduction.
What is the role of malate dehydrogenase in cellular metabolism?
Malate dehydrogenase (MDH) (EC 1.1.1.37) catalyzes the conversion of oxaloacetate and malate. This reaction is important in cellular metabolism, and it is coupled with easily detectable cofactor oxidation/reduction. It is a rather ubiquitous enzyme, for which several isoforms have been identified, d …
What is the structure of MDH?
Despite the low amino acids sequence identity their 3-dimensional structures are very similar. MDH is a group of multimeric enzymes consisting of identical subunits usually organized as either dimer or tetramers with subunit molecular weights of 30-35 kDa.
Can nucleotide binding be altered?
The nucleotide binding characteristics can be altered by a single amino acid change. Multiple amino acid sequence alignments of MDH show that there is a low degree of primary structural similarity, apart from several positions crucial for catalysis, cofactor binding and the subunit interface.
What are the substrates of malate dehydrogenase?
Thus, the two substrates of this enzyme are (S)-malate and NAD +, whereas its three products are pyruvate, CO 2, and NADH.
What is the name of the enzyme that catalyzes the chemical reaction?
Malate dehydrogenase (decarboxylating) Malate dehydrogenase (decarboxylating) ( EC 1.1.1.39) or NAD-malic enzyme (NAD-ME) is an enzyme that catalyzes the chemical reaction. Thus, the two substrates of this enzyme are (S)-malate and NAD +, whereas its three products are pyruvate, CO 2, and NADH. Malate is oxidized to pyruvate and CO 2, ...
What enzymes are involved in pyruvate metabolism?
This enzyme participates in pyruvate metabolism and carbon fixation. NAD-malic enzyme is one of three decarboxylation enzymes used in the inorganic carbon concentrating mechanisms of C 4 and CAM plants. The others are NADP-malic enzyme and PEP carboxykinase.
What are the substrates of a pyruvate enzyme?
Thus, the two substrates of this enzyme are (S)-malate and NAD +, whereas its three products are pyruvate, CO 2, and NADH. Malate is oxidized to pyruvate and CO 2, and NAD + is reduced to NADH.
What is the role of malate dehydrogenase in the Krebs cycle?
It is responsible for oxidizing 4 carbon malate into 4 carbon oxaloacetate. Malate Dehydrogenase usually requires the use of a NAD+ to act as an electron acceptor when malate is oxidized. There are multiple isoforms of this enzyme, and the primary isoform is located in the mitochondrial matrix.
What enzyme is responsible for oxidation of malate to oxaloacetate?
Malate dehydrogenase (MDH) is an enzyme that reversibly catalyzes the oxidation of malate to oxaloacetate using the reduction of NAD to NADH.
Where does adenylated amino acid go?
Adenylated amino acid remains tightly bound to the aminoacyl tRNA synthetase enzyme and tRNA, specific for the enzyme, enters the active site. This would result in the transfer of the amino acid (carboxyl group) to the 3′ or 2′ OH end (5′CCA3′/2′) of the tRNA and release of AMP.
What isomerases can catalyze backward reaction?
Take the example of isomerases. Suppose, triose phosphate isomerases. They convert dehydroxy acetone phosphate (DHAP) to 3-phospho glyceraldehyde (3-PGAld). They can also catalyse the backward reaction (i.e. convertion of 3-PGAld to DHAP). Although the nature of reaction is same (i. e. Isomerisation) but they are two different reactions.
Why are enzymes so picky?
Enzymes are really picky - they usually only catalyze one specific reaction. This is because, for the enzyme to work, the substrate has to fit into its active site. If the substrate doesn’t match the enzyme’s active site, then the reaction won’t be catalyzed.
Is LDH measured in clinical labs?
There is very little use in the clinical measurement of LDH. In my last lab the only real requests were in abdominal pain cases. A protocol is in use for grading acute pancreatitis which includes LDH measurement. Otherwise the only measurements of LDH were EQAS (External Quality Assurance Samples). The US equivalent of this would be PT (Proficiency Testing).
What is the role of malate dehydrogenase in the tricarboxylic acid cycle?
Malate dehydrogenase (MDH), a key enzyme in the tricarboxylic acid cycle (TCA), plays important metabolic roles in aerobic energy producing pathways and in malate shuttle (Figure 1) [5].
What enzymes have bands?
Bands were revealed for the enzyme systems esterase (EST), according to Scandalios (1969), alcohol dehydrogenase (ADH), phosphoglucose isomerase (PGI), malate dehydrogenase (MDH) and malic enzyme (ME), described by Alfenas (1991), and alpha amylase ( [alpha] - AMY), according to Alfenas (1998).
What is the function of malate dehydrogenase?
Malate dehydrogenase (MDH) (EC 1.1.1.37) catalyzes the conversion of oxaloacetate and malate. This reaction is important in cellular metabolism, and it is coupled with easily detectable cofactor oxidation/reduction.
Which enzymes are closely related to MDH?
The MDH amino acid sequences show divergence into two main phylogenetic groups of closely related enzymes - cytoplasmic and mitochondrial M DHs. Some of the most distanly related isoenzymes of MDH are found compartmentalized in different subcellular organelles of the same cell types (Goward and Nicholls 1994).
What is the function of Musrati 202?
202 Musrati et al. boxylated via NADP-malic enzyme, it acts as a carrier of reducing power as well as CO2. The NADPH formed is directly utilised for phosphoglycerate reduction. The encoded amino acid sequence of maize predicts that NADP-MDH is syn thesized as a preprotein of 432 amino acids and processed into a mature protein of 375 amino acids with removal of a 57 amino acids long transit peptide. The sorghum enzyme is synthesized as a precursor of 429 amino acids and gets imported into the chloroplast where it is processed to a mature subunit of 389 amino acids. De spite the lack of sequence similarities to other chloroplast transit peptides the extra sequence shows the common features : it is rich in the hydroxylated amino acids serine and threonine (14%), it is also rich in small hydrophobic amino acids such as alanine and valine (28%), it shows net positive charge (8 arginines, 1 lysine), and is generally deficient in acidic amino acids (2 aspartates). The maize enzyme is similar to other MDHs in regions related to enzymatic function (Wilks et al. 1988). The similarity of the C3 and C4 forms of NADP-MDH suggests that genes for C4 enzymes may have been recruited from existing genes encoding C3 enzymes (Gietl 1992). Eubacterial MDH Escherichia coli malate dehydrogenase (eMDH) is a homodimer and it is com prised of 312 amino acid residues per subunit (Sutherland and McAlister-Henn 1985; McAlister-Henn et al. 1987). The structures of cytosolic and mitochondrial MDHs from porcine heart have been determined (Roderick and Banaszak 1986; Birktoft et al. 1989a) and a comparative study between these enzymes and eMDH demonstrated that eMDH has 58% sequence identity with mMDH, but only about 20% identity with cMDH (Hall et al. 1992; Gleason et al. 1994). In the crystalline structures of 2 prokaryotic and 2 eukaryotic forms, the sub- unit interfaces are conformationally homologous. To determine whether or not the quaternary structure of MDH is linked to catalytic activity, mutant forms of the enzyme from E. coli have been constructed. Utilizing the high-resolution structure of E. coli MDH, the dimer interface was analyzed critically for side chains that were spatially constricted and needed for electrostatic interactions. Two such residues were found, D45 and S226. At the nearest point in the homodimer, they were found in different subunits, hydrogen bonded across the interface, and did not interact with any catalytic residues. Each residue was mutated to a tyrosine, which should disrupt the interface because of its large size. All mutants were cloned and purified to homogeneity. Gel fitration of the mutants showed that D45Y and D45Y/S226Y were both monomers, whereas the S226Y mutant remained a dimer. The monomeric D45Y and D45/S226Y mutants had 14,000 and 17,500-times less activity, respec tively, than the native enzyme. The dimeric S226Y had only 1.4-times less specific activity.
How many bases are in cMDH?
198 Musrati et al. number of charged residues. There are 41 basis residues (31 lysines + 10 arginines) and 43 acidic residues (25 aspartates + 18 glutamates) in cMDH. In mMDH there are 25 lysines and 8 arginines for total of 33 basic residues, and 13 aspartates and 16 glutamates for a total of 29 acidic groups (Birktoft et al. 1989a). The sequence identity between cMDH and mMDH is relatively low, being of the order of about 20-25%. The best fit of the molecular structure of cMDH to that of lactate dehydrogenase has been obtained by the least square method. This similarity between the dimeric cMDH and the tetrameric LDH reported earlier by Rao and Rossmann (1973), particularly in the nucleotide binding domain, has been confirmed. The active sites of these two enzymes contain similarly oriented His-Asp pairs linked by a hydrogen bond which may function as a proton relay system during catalysis. This pair could also provide an explanation for the relatively stronger binding by cMDH and LDH of NADH versus NAD. Peroxisomal and glyoxysomal MDH Peroxisomes are organelles present in almost all eukaryotic cells. Like the mitochon drion, the peroxisome is a major site of oxygen utilization. In fact, the peroxisome is thought by some to represent the vestige of an ancient organelle that carried out all of the oxygen metabolism of primitive preeukaryotic cells when oxygen en tered the atmosphere. Most peroxisomes catalyze the breakdown of fatty acids to acetyl-CoA using a special H2 02-producing enzyme. The acetyl-CoA produced can be transported via the cytosol to the mitochondria to feed the citric acid cycle, or it can be used for biosynthetic reactions elsewhere. Two very different types of peroxisomes have been extensively studied in plants. One type is present in leaves, where it catalyzes the oxidation of a side product of the reaction that fixes CO2 in carbohydrate (photorespiration). A very different type of peroxisome is present in germinating seeds, where it serves to con vert the fatty acids stored in seed lipids into sugars needed for the production of the materials of the young plant. Because this is accomplished by a series of reactions known as the glyoxylate cycle, these peroxisomes are also called glyoxysomes. Metabolism of glycolate carbon occurs sequentially in three organelles, the per oxisomes, the mitochondria and the chloroplasts (Lorimer and Andrews 1980). In the peroxisome glycolate is oxidised to glyoxylate and then transaminated to glycine with either glutamate or serine (Rehfeld and Tolbert 1972). Glycine then stays in the peroxisome and is oxidised to ammonia, CO2 and serine in the mitochondrion. Serine is converted to glycerate by serine:glyoxylate aminotransferase and hydrox- ypyruvate reductase in the peroxisome, and in this form photorespiratory carbon returns to the chloroplast. Glycerate is phosphorylated to phosphoglycerate by glycerate kinase, and can reenter the photosynthetic cycle. Photorespiration is an example for the tight cooperation of metabolic pathways
Where is NADP-MDH found?
200 Musrati et al. ies, the mitochondria and the cytoplasm, an NADP-dependent form of the enzyme is found in the chloroplasts in higher plants. NADP-dependent MDH (NADP- MDH) (EC. 1.1.1.82) is involved in the C4dicarboxylic acid cycle responsible for the primary fixation and transfer of CO2 in C4 plants. It is also present in C3 type plants where it is implicated in chloroplast shuttle mechanisms which might help export reducing power. NADPH-MDH is a model enzyme in that its activity is strictly controlled by light (Johnson and Hatch 1970). Indeed, NADP-MDH is inactive in the dark and gets activated by light via the ferredoxin - thioredoxin system (Wolosiuk et al. 1980). NADP-MDH resembles the non-regulatory NAD-MDH, except for two se quence additions, one N-terminal and one C-terminal. Due to the presence of both N- and C-terminal extensions chloroplastic MDHs exhibit larger molecular masses, and they represent crucial parts of the protein for redox regulation. Several cysteine residues are located in the NADP-MDH polypeptide, and all are specific to this chloroplastic redox regulatory isoform. Two of these are located in the N-terminal extension sequence and one in the C-terminal extension. Chemical derivatization followed by sequence analysis in sorghum (Decottignies et al. 1988) has shown that there is a light-dependent reducible disulphide bridge present in the N-terminal extension. Site-directed mutagenesis has indeed confirmed that there is a thioredoxin-reducible disulphide bridge involving these two residues (Issakidis et al. 1992), but has also shown that there is a second disulphide needed for regulation. Additional site-directed mutagenesis experiments have succeeded in creating a redox insensitive NADP-MDH, by mutation of the N-terminal disulphide, together with either or both of the most C-terminal cysteines (C377 and/or C365 in sorghum), indicating that these cysteines could constitute the second regulatory disulphide (Issakidis et al. 1993; 1994). There is experimental evidence that the disulfide bridge involving the C-terminal residues is shielding access to the catalytic residues, and that the N-terminal end is involved in the slow conformational change of the active site needed for activation (Issakidis et al. 1996). On the basis of the results of mutagenesis experiments and three-dimensional structure modeling of the chloroplastic isoenzyme, Issakidis et al. (1994) proposed a model for the mechanism of activation of NADP-MDH (Fig. 2). Chloroplast NADP-MDH possesses a His-Asp pair at the active site which probably forms a proton relay system. The involvement of such a His/Asp pair in catalysis has already been described for LDH and NAD-MDH. It is acting as an acid in the reduction of the keto-acid and as a base in the oxidation of the hydroxy-acid (Birktoft and Banaszak 1983). So, His-229 and Asp-201 play a crucial role in the catalytic mechanism of chloroplastic NADP-MDHs (Lemaire et al. 1996). Sequence data of NADP-MDH from pea (Reng et al. 1993), sorghum (Cretin et al. 1990), maize (Metzler et al. 1989) and ice-plant are available.
Where is MDH synthesized?
Both forms of MDH are synthesized in the cy toplasm, where cMDH remains after acetylation of the N-terminal residue. mMDH is synthesized with an N-terminal extension residue and is subsequently imported into the mitochondrial matrix (Chien and Freeman 1984).
Where is MDH found in eukaryotic tissue?
MDH in eukary otic organisms is found in microbodies such as glyoxysomes and peroxisomes, in the mitochondria, in the cytoplasm and in chloroplasts.
What is magnesium malate made of?
Magnesium malate is a compound made by combining magnesium with malic acid.
What hormone is responsible for transporting sugar from bloodstream into tissues?
Insulin is the hormone responsible for transporting sugar from your bloodstream into your tissues. Increasing insulin sensitivity can help your body use this important hormone more efficiently to keep your blood sugar levels in check ( 11.
Can magnesium malate be taken with magnesium malate?
Common uses. Research shows that the majority of adults in the United States consume less magnesium than is recommended ( 3 ). You can take magnesium malate to help increase your intake of magnesium. This can help prevent magnesium deficiency if you’re not getting enough in your diet. Many people also use magnesium supplements to help prevent ...
Can magnesium malate cause diarrhea?
Magnesium malate may cause side effects like nausea, diarrhea, and stomach cramps. It can also be toxic in very high doses and may interfere with certain types of medications.
Does magnesium malate help with fibromyalgia?
Fibromyalgia is a chronic condition that causes muscle pain and tenderness throughout the body ( 18 ). Some research suggests magnesium malate could help reduce its symptoms. One study in 80 women found that blood levels of magnesium tended to be lower in those with fibromyalgia.
What is malate in the TCA cycle?
Malate is an essential intermediate of the Tricarboxylic Acid (TCA) Cycle, which generates two-thirds of the body’s energy by utilising fats and carbohydrates consumed through the diet. In the TCA cycle, malate is dehydrogenised (stripped of its hydrogen atoms), which, in the process, generates NADH2. This co-enzyme can then go on to produce adenosine triphosphate (ATP), which is the energy needed for muscle contraction.5
What is the name of the enzyme that produces NADH2?
Malate . Malate is an essential intermediate of the Tricarboxylic Acid (TCA) Cycle, which generates two-thirds of the body’s energy by utilising fats and carbohydrates consumed through the diet. In the TCA cycle, malate is dehydrogenised (stripped of its hydrogen atoms), which, in the process, generates NADH2.
What is Citrulline Malate?
Citrulline malate (CM) is an amino acid compound that has gained attention due to its reported ability to delay the onset of fatigue during intense exercise. That’s why you’ll often find CM in pre-workout supplements. Let’s take a closer look at its components…
Does citrulline malate help with aerobic exercise?
Not only does citrulline malate have benefits for power related exercise, it has also shown benefits in aerobic exercise.10. A few years ago, a research group put this to the test using a high-intensity cycling protocol, after participants had consumed either citrulline malate or placebo for the previous 7 days.
Does citrulline malate increase reps?
Similar effects have been observed in women, with citrulline malate supplementation increasing the number of reps completed of various upper- and lower- body resistance exercises, compared to the placebo .
Does citrulline malate cause fatigue?
Citrulline malate can delay muscle fatigue. During one cross-over study, resistance-trained men completed 5 sets of leg press, hack squat, and leg extension at 60% of one-rep max (the maximum weight you can lift for one repetition) until failure.
Is citrulline malate an ergogenic?
Take Home Message. All in all, citrulline malate looks like it has great potential as an ergogenic aid for both resistance and endurance performance. The citrulline component will help to boost nitric oxide production and shift unwanted ammonia, reducing muscular fatigue in the process.

Safety
- It is expected that none of these abovementioned parameters on its own will be the universal safety parameter for liver; instead, a combination of leakage and function markers may be added to a panel of tests showing adequate sensitivity and specificity for most non-clinical and clinical …
Structure
- Several isoforms of MDH have been identified, differing in their subcellular localization and their specificity to the coenzyme NAD or NADP. In eukaryotic cells, at least two forms of the enzyme can be found. One isoform is a principal enzyme of the citric acid cycle operating within mitochondria [68]. The other is found in the cytosol where it participates in the malate/aspartate …
Synthesis
- Malate dehydrogenase (cytoplasmic). Oxidation of malate in the cytoplasm regenerates OAA and nicotinamide adenine dinucleotide (NADH). The latter is needed at reaction step 8 (glyceraldehyde-3-phosphate dehydrogenase; see below). Phosphoenolpyruvate carboxykinase (PEPCK). Decarboxylation of OAA to produce PEP is accompanied by phosphorylation using gua…
Funding
- MDH is supported by grants from the Medical Research Council of the United Kingdom and the Wellcome Trust. GBB was supported by grants from the Department of Veterans Affairs and the National Cancer Institute, USA, as well as the NCI-supported core facilities of the Huntsman Cancer Institute. We thank Drs. T. J. Torphy, R. J. Owen, M. Perry, J. Souness, K. Jarnagin, E. R. S…
Clinical significance
- The enzyme MDH is involved in the Krebs cycle, catalyzing the reversible transformation of malate into oxaloacetate utilizing NAD+. It is also a leakage marker released into the serum after tissue damage, being highly abundant in liver followed by heart, skeletal muscle, and brain. This enzyme is localized in the mitochondria (10%) and in the cytoplasm (90%) (Marrer and Dieterle, 2010). M…
Mechanism
- The interaction between CS and mMDH has been demonstrated using a number of other physical techniques. Using fluorescently labeled proteins, it was shown that with CS and mMDH a change of anisotropy was observed. A KDISS of 10 6 M was calculated using this technique (Tompa et al., 1987). Beeckmans et al. (1989) used an affinity electrophoresis technique to show specific inter…
Interactions
- The technique of precipitation with PEG was employed to show specific interaction between CS and mitochondrial aconitase (mACO) and between mACO and NAD-specific isocitrate dehydrogenase (NAD-ICDH) (Tyiska et al., 1986; Srere, 1987). Using one or more of these techniques outlined above, we know that NAD-ICDH interacts with α-KGDC (Porpaczy et al., 1987…
Overview
Malate dehydrogenase (EC 1.1.1.37) (MDH) is an enzyme that reversibly catalyzes the oxidation of malate to oxaloacetate using the reduction of NAD to NADH. This reaction is part of many metabolic pathways, including the citric acid cycle. Other malate dehydrogenases, which have other EC numbers and catalyze other reactions oxidizing malate, have qualified names like malate dehydrogen…
Isozymes
Several isozymes of malate dehydrogenase exist. There are two main isoforms in eukaryotic cells. One is found in the mitochondrial matrix, participating as a key enzyme in the citric acid cycle that catalyzes the oxidation of malate. The other is found in the cytoplasm, assisting the malate-aspartate shuttle with exchanging reducing equivalents so that malate can pass through the mitochondrial membrane to be transformed into oxaloacetate for further cellular processes.
Evolution and structure
In most organisms, malate dehydrogenase (MDH) exists as a homodimeric molecule and is closely related to lactate dehydrogenase (LDH) in structure. It is a large protein molecule with subunits weighing between 30 and 35 kDa. Based on the amino acid sequences, it seems that MDH has diverged into two main phylogenetic groups that closely resemble either mitochondrial isozymes or cytoplasmic/chloroplast isozymes. Because the sequence identity of malate dehydrogenase i…
Mechanism
The active site of malate dehydrogenase is a hydrophobic cavity within the protein complex that has specific binding sites for the substrate and its coenzyme, NAD . In its active state, MDH undergoes a conformational change that encloses the substrate to minimize solvent exposure and to position key residues in closer proximity to the substrate. The three residues in particular t…
Function
Malate dehydrogenases catalyzes the interconversion of malate to oxaloacetate. In the citric acid cycle, malate dehydrogenase is responsible for catalyzing the regeneration of oxaloacetate This reaction occurs through the oxidation of hydroxyl group on malate and reduction of NAD . The mechanism of the transfer of the hydride ion to NAD is carried out in a similar mechanism …
Effect of pH on catalytic activity
Additionally, pH levels control specificity of substrate binding by malate dehydrogenase due to proton transfer in the catalytic mechanism. A histidine moiety with a pK value of 7.5 has been suggested to play a role in the pH-dependency of the enzyme. Studies have indicated that the binding of the enol form oxaloacetate with the malate dehydrogenase:NADH complex forms much more rapidly at higher pH values. Additionally, L-malate binding to malate dehydrogenase is pro…
Allosteric regulation
Because malate dehydrogenase is closely tied to the citric acid cycle, studies have proposed and experimentally demonstrated that citrate is an allosteric regulator of malate dehydrogenase depending on the concentrations of L-malate and NAD . This may be due to deviations observed in the kinetic behavior of malate dehydrogenase at high oxaloacetate and L-malate concentrations. Experiments have shown that Citrate can both allosterically activate and inhibit t…
Interactive pathway map
Click on genes, proteins and metabolites below to link to respective articles.