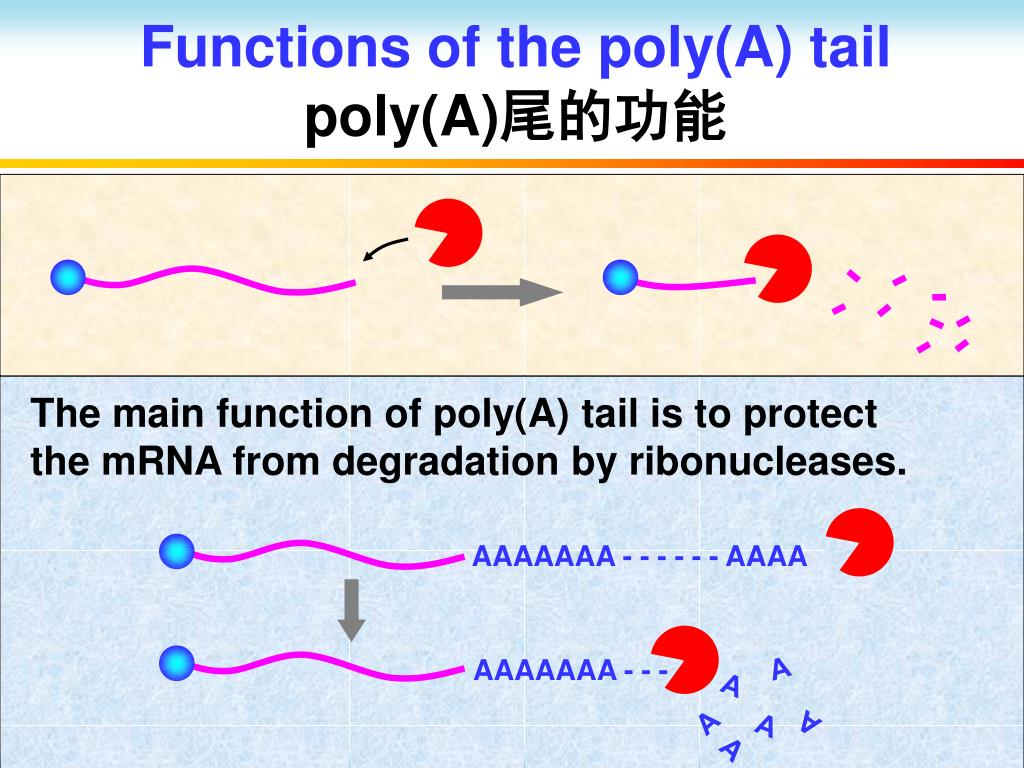
How does the polyadenylation signal work? Function. In nuclear polyadenylation, a poly (A) tail is added to an RNA at the end of transcription. On mRNAs, the poly (A) tail protects the mRNA molecule from enzymatic degradation in the cytoplasm and aids in transcription termination, export of the mRNA from the nucleus, and translation.
What is the function of polyadenylation?
Polyadenylation is the post-transcriptional additional of multiple adenine (A) nucleotides to the tail of a messenger RNA transcript. The purpose and mechanism of polyadenylation vary across cell types, but polyadenylation generally serves to promote transcript longevity in eukaryotes and promote transcript degradation in prokaryotes.
What is the cleavage site associated with a polyadenylation signal?
The cleavage site associated with a polyadenylation signal can vary up to some 50 nucleotides. When the RNA is cleaved, polyadenylation starts, catalysed by polyadenylate polymerase. Polyadenylate polymerase builds the poly (A) tail by adding adenosine monophosphate units from adenosine triphosphate to the RNA, cleaving off pyrophosphate.
What is polyadenylation of mRNAs?
mRNA polyadenylation is an essential step for the maturation of almost all eukaryotic mRNAs and is tightly coupled with termination of transcription in defining the 3′-end of genes. A polyadenylation signal (AAUAAA) nearby the 3′-end of pre-mRNA is required for poly(A) synthesis.
What is the role of polyadenylation in RNA degradation in Escherichia coli?
Polyadenylation promotes degradation of 3'-structured RNA by the Escherichia colimRNA degradosome in vitro. J Biol Chem. 1999;274:4009–4016. [PubMed: 9933592] 67. Coburn GA, Miao X, Briant DJ. et al. Reconstitution of a minimal RNA degradosome demonstrates functional coordination between a 3' exonuclease and a DEAD-box RNA helicase.

What is the polyadenylation signal?
The polyadenylation signal is the categorization motif documented by a compound called ribonucleic acid (RNA) cleavage that varies amongst the collections of eukaryotes. Most humanoid polyadenylation sites comprise the AAUAAA order, but this order is less communal in floras as well as fungi. The ribonucleic acid is sliced right afterward transcript, as CstF also offers to RNA polymerase II.
Why is the poly-A tail important?
The poly-A tail is an important part of the mRNA because it is the long chain of a nucleotide, known as adenosine attached to the mRNA to increase its stability. The process by which enzymes add about a hundred to two hundred adenine to the 3’ end of the mRNA is polyadenylation. Along with providing stability, the poly-A tail also helps the mRNA to form the proteins by exporting it out of the nucleus towards the cytoplasm to be read by the ribosome for the translation process.
How does polyadenylation affect sequencing?
Polyadenylation of footprint fragments introduces difficulties in mapping a sequence to its genomic origin. Ribosome footprint fragments are shorter than the 36 nt sequencing read length provided by the Genome Analyzer, so some poly-A sequence will be present at the end of most sequencing reads. It is not always possible to know how many terminal As in a sequencing read were added during polyadenylation and how many were derived from the RNA fragment ( Fig. 6.7 ). Furthermore, sequencing errors are more likely near the end of the read, making it more difficult to identify the exact extent of the terminal poly-A region. One approach that avoids these difficulties is to align a “seed” from the beginning of each sequencing read to the reference sequence and then to determine the full extent of the alignment. The Bowtie short sequence alignment program is fast, robust, and well-suited to performing these seed alignments ( Langmead et al., 2009 ). Furthermore, it reports alignments to targets in the order in which they appear in the query file, which facilitates postprocessing of seed alignments. Finally, it is capable of reporting multiple alignments as well as reporting on degenerate and unaligned query sequences.
What is polyadenylation in mRNA?
Polyadenylation is an obligatory feature of nuclear pre-mRNA processing and is catalyzed by a “cano nical” poly (A) polymerase, that is, an orthologue of Saccharomyces cerevisiae Pap1p. From: The Enzymes, 2012. Download as PDF.
What is cytoplasmic polyadenylation?
Cytoplasmic polyadenylation is another mechanism by which mRNA translation is regulated upon synaptic stimulation (Steward and Schuman, 2001 ). The molecular mechanisms underlying cytoplasmic polyadenylation have been extensively characterized in Xenopus oocytes, where a subset of dormant mRNAs have small poly (A) tails (usually ∼20–40 nucleotides in length) ( Mendez and Richter, 2001 ). In response to developmental cues, the poly (A) tails of the dormant mRNAs are lengthened, and translation ensues. Polyadenylation is controlled by two cis elements in the 3′UTR of the mRNA: the cytoplasmic polyadenylation element (CPE, sequence of UUUUUAU) and the hexanucleotide AAUAAA, which is also important for nuclear pre-mRNA cleavage and polyadenylation ( Fox et al., 1989; McGrew et al., 1989 ). Cytoplasmic polyadenylation element (CPE) binding factor (CPEB), which contains an RNA recognition motif and zinc finger domain, binds with strong affinity with the CPE ( Hake and Richter, 1994; Hake et al., 1998) and interacts with a number of other regulatory factors. These factors include (1) symplekin, a scaffold-like protein; (2) cleavage and polyadenylation specificity factor (CPSF), a group of four proteins that binds the hexanucleotide AAUAAA; and (3) Gld2 ( Barnard et al., 2004 ), an unusual poly (A) polymerase first discovered in yeast and Caenorhabditis elegans ( Read et al., 2002; Saitoh et al., 2002; Wang et al., 2002 ). Polyadenylation is initiated when aurora (Eg2) phosphorylates CPEB ( Mendez et al., 2000a ), which induces CPEB to interact and possibly stabilize CPSF on the hexanucleotide AAUAAA ( Mendez et al., 2000b ). This process is likely necessary for the recruitment of poly (A) polymerase. One mechanism by which polyadenylation is believed to stimulate translation involves maskin, a protein that interacts with both CPEB and eIF4E ( Stebbins-Boaz et al., 1999 ). The maskin–eIF4E interaction inhibits translation by precluding the eIF4E–eIF4G interaction and formation of the eIF4F complex and ribosomal recruitment to the mRNA ( Cao and Richter, 2002 ). Polyadenylation leads to the recruitment of the poly (A)-binding protein, which binds to eIF4G ( Wakiyama et al., 2000 ), a process that enhances the eIF4G–eIF4E association and promotes the dissociation of maskin from eIF4E ( Cao and Richter, 2002 ).
What is the name of the enzyme that catalyzes the polyadenylation of mtRNA?
In contrast to bacteria and some eukaryotes where polyadenylation is catalyzed by poly (A) polymerases (PAPs) or PNPase, human mtRNA is polyadenylated by mitochondria-specific PAP.
What is the polyA tail?
The polyA tail is a long chain of adenine nucleotides that is added to a mRNA molecule during RNA processing. The polyA tail makes the RNA molecule more stable and prevents its degradation and allows the mature mRNA molecule to be exported from the nucleus and translated into a protein by ribosomes in the cytoplasm. A comparison of different polyA signals, including the mouse α-globin polyA, the bovine growth hormone polyA, and a synthetic polyA showed that by utilizing different polyA sequences expression could be modulated over a 7-fold range ( Wu et al., 2008 ).
What are the ambiguities in alignments of sequencing reads due to polyadenylation?
A sample genomic sequence from the start of the GCN4 gene is shown along with five possible footprinting products that differ only in the position of their 3′ terminus. Polyadenylation adds poly- (A) tails, shown in gray, but sequencing cannot distinguish which nucleotides occurred in the original RNA fragment and which were added. The middle three sequencing reads are all identical, whereas the 3′ termini of the top and bottom read can be uniquely distinguished. A table of inferred reference alignment lengths lmin and lmax is given for each sequencing read.
What is the function of hPNPaseold-35?
Figure 5.9. Functions of hPNPaseold-35. hPNPaseold-35 is transcriptionally induced by the JAK/STAT pathway through IFN-β produced by the cell after viral infection, exposure to tumor cells , senescence-inducing signals or terminal-differentiation promo ting signals. Next, hPNPase old-35 is imported to the mitochondrial inter membrane space (IMS) by an YME-mediated mechanism or alternatively mobilized through an unknown mechanism to the cytoplasm where it causes growth arrest or apoptosis by targeting c-myc mRNA and miR-221 or activating PKR, respectively. Exposure to ionizing radiation inactivates hPNPase old-35 by an EGFR-mediated mechanism, causing c-myc mRNA upregualtion and increasing radioresistance of cancer cells. In the mitochondria, hPNPase old-35 maintains mitochondrial homeostasis, aids in RNA import, takes part in mtRNA processing/degradation events, and increases ROS production that results in events leading to chronic inflammation.
What is polyadenylation in RNA?
Polyadenylation is a postranscriptional modification of RNA found in all cells and in organelles. In bacteria, a small fraction of RNA harbors oligo(A) tails which are mostly shorter than 20 As. Poly(A) polymerase I of Escherichia coli can adenylate mRNAs, and small RNA regulators originating from the chromosome, from plasmids and from bacteriophages and also precursors and mature forms of tRNAs and rRNAs which have an accessible 3' extremity. The model of poly(A) metabolism presented in this chapter proposes that the ratio of adenylated to nonadenylated RNAs and the length of oligo(A) tails represent the equilibrium of the antagonistic activities of poly(A) polymerase I, which synthesizes poly(A) at the 3' end of all RNAs, and of exoribonucleases which shorten or completely remove the tails. Poly(A) tails provide toeholds where polynucleotide phosphorylase can initiate exonucleolytic degradation of tightly folded RNAs protected from exoribonucleases by 3' stable secondary structures. Polyadenylation promotes degradation of mRNA fragments and controls the intracellular concentration of regulatory RNAs.
What gene reduces plasmid copy number of pBR322?
Lopilato J, Bortner S, Beckwith J. Mutations in a new chromosomal gene of Escherichia coliK-12, pcnB, reduce plasmid copy number of pBR322 and its derivatives. Mol Gen Genet. 1986;205:285–290. [PubMed: 3100913]
What is the function of polyadenylation?
In eukaryotic organisms’ polyadenylation is considered as a part of process which produces mature messenger RNA for the process of translation. In many of the bacteria’s the poly (a) tail promotes degradation in the mRNA. Which therefore forms a part of the large processor in the gene expression. The poly (A) tail is very much important for ...
What is polyadenylation in RNA?
Features of Polyadenylation. Polyadenylation is a process of addition of poly (A) tail to transcript RNA, typically a messenger RNA (mRNA). The poly (A) tail contains multiple of adenosine monophosphate, in other terms it can also be said that it is a stretch of RNA which contains only adenine bases. In eukaryotic organisms’ polyadenylation is ...
How many nucleotides does a polyadenylation tail have?
When the poly (A) tail reaches an approximate of 250 nucleotides long it cannot bind with the CPSF and thus the polyadenylation stops determining the length of the poly (A) tail. CPSF is in contact with the RNA polymerase II thus allowing it to rise a signal to the polymerase to end the process of transcription.
What is polyadenylic acid tail?
The term poly (A) denotes polyadenylic acid tail which reflects the RNA nucleotides for a base Adenine denoted by a letter A, and the other bases where G is noted for Guanine and C is noted for cytosine and U is noted as Uracil. RNA is produced from the DNA during the transcribing process.
Which mRNA molecules are polyadenylated?
The mRNA molecules which is present in both prokaryotes and eukaryotes have the polyadenylated 3’ends, along with the prokaryotic poly (A) tails are generally shorter and less mRNA molecules polyadenylated.
Where does the polyadenylation process occur?
Polyadenylation process initiates in the nucleus of the eukaryotes which works in the RNA polymerase II, as a precursor mRNA, Where the 3’ part of the newly produced RNA and the polyadenylates are resultant of this cleavage process.
Why is the poly tail important?
The poly (A) tail is very much important for the nuclear export, translation and in maintain the stability of mRNA. The tail is shortened upon time until it becomes short enough when the mRNA is enzymatically degraded.
What is the purpose of polyadenylation?
The purpose and mechanism of polyadenylation vary across cell types, but polyadenylation generally serves to promote transcript longevity in eukaryotes and promote transcript degradation in prokaryotes.
What are termination and polyadenylation?
Terminators are found downstream of the gene to be transcribed, and typically occur directly after any 3’ regulatory elements, such as the polyadenylation or poly (A) signal. While many studies focus on promoter strength as a determinant of gene expression levels, the terminator also plays an important role in RNA processing and contributes to variability in RNA half-life, and ultimately gene-expression.
What is a plasmid 101?
Plasmids 101: Terminators and PolyA signals. Plasmids designed to express genes in a given host cell type are generally broken down into two broad categories, prokaryotic or eukaryotic, based on the functional elements they contain. Plasmid DNA in both prokaryotic and eukaryotic systems must be transcribed into RNA, ...
Which signal directs cleavage at the cleave signal?
Figure 2: The conserved eukaryotic polyadenylation signal directs cleavage at the cleave signal and addition of a poly-A tail to the mRNA transcript. By Arunreginald at en.wikipedia, CC BY-SA 3.0, https://commons.wikimedia.org/w/index.php?curid=17554393
Do prokaryotes have poly tails?
Although mostly thought of as a eukaryotic-specific process, prokaryotes also add poly (A) tails to certain RNA s. Unlike the eukaryotic mechanism which requires a consensus sequence for the addition of a poly (A) tail, the addition of a poly (A) tail on a prokaryotic transcript is non-specific and can be added to any accessible 3' end. The presence of the poly (A) tail targets the RNA to the degradosome, which contains enzymes that cut RNA not protected by secondary structure. It is thought that poly (A)s are used to control the cellular concentration of regulatory RNAs and may additionally act as a quality control mechanism to rid the cell of mis-folded RNAs.
What sequence is required for polyadenylation?
Although the AAUAAA sequence was shown to be absolutely required for mRNA 3′-end polyadenylation, other sequence elements were shown to be necessary to fully reconstitute a functional PAS. In particular, the GU-rich sequence (or downstream sequence element, DSE) present just past the mRNA 3′ end in the immediate gene 3′ flanking region was shown to enhance 3′-end formation ( Gil and Proudfoot 1984, 1987; McLauchlan et al. 1985 ). Similarly, the sequence immediately upstream of AAUAAA (upstream sequence element, USE) ( Carswell and Alwine 1989; DeZazzo et al. 1991; Valsamakis et al. 1991; Moreira et al. 1995; Brackenridge and Proudfoot 2000; Venkataraman et al. 2005; Danckwardt et al. 2007) can in some cases act as an enhancing element for 3′-end processing efficiency. Finally, the actual nucleotides at the site of 3′-end cleavage can also influence the efficiency of this process ( Chen et al. 1995 ). This proved to be medically relevant for the human prothrombin gene, which normally carries a CG dinucleotide sequence just 5′ to the cleavage position. In ∼1% of Caucasians (myself included), this sequence is mutated to CA, causing a mild thrombophilia phenotype. This is due to a twofold increase in prothrombin gene expression as a result of the acquisition of a more efficient PAS ( Gehring et al. 2001; Danckwardt et al. 2008 ). These defining experiments for mRNA PAS were focused on mammalian mRNA. However, other eukaryotic model systems revealed that the mammalian pattern of USE-AAUAAA-DSE ( Fig. 2B) was generally conserved across eukaryotes, even though in budding yeast greater variation in these cis elements appears to be tolerated ( Graber et al. 1999 ). One notable feature of PAS is that, depending on exactly how the AAUAAA and DSE signals are positioned and defined, the actual site of poly (A) addition can vary by several to tens of nucleotides ( Zhang et al. 1986; Sheets et al. 1990; Tian et al. 2005 ). This is especially evident in yeast PAS, which, perhaps due to lower sequence conservation, do not accurately define the position of polyadenylation ( Zhao et al. 1999 ). This is in marked contrast to pre-mRNA splicing, which must always occur at a precise nucleotide position to maintain the correct reading frame of the mRNA for translation.
What is the Northern Blotting technique?
The characterization of particular eukaryotic mRNAs has often relied on the long-established Northern blotting technique, which provides a gel fractionation image of a specific gene's mRNA output, giving information relating to both mRNA size and quantity. This technique—unlike its modern replacement of quantitative (real-time) PCR-amplified cDNA (qRT–PCR)—allows visualization of the complete set of mRNA isoforms generated from a particular gene. Smaller genes often yield only one specific mRNA, which is the norm for simpler eukaryotes such as yeast. However, in higher eukaryotes, especially mammals, most of these larger and more complex genes (with multiple exons) generate multiple mRNA isoforms. These are frequently caused by complex alternative splicing patterns. Alternative splicing is now appreciated to regulate both the nature and complexity of mammalian proteomes and, as such, reflects a key aspect of the regulation of gene expression ( Black 2003; Johnson et al. 2003; Wang et al. 2008; Chen and Manley 2009 ). However, a significant part of mRNA size variation derives not from alternative splicing, but rather from alternative PAS selection. Thus, it is calculated that well over half of all mRNAs have variable PAS selection, meaning that they will possess mRNA isoforms differing by the extent of their 3′ UTRs ( Edwalds-Gilbert et al. 1997; Tian et al. 2005 ). Since mRNA 3′-end processing occurs cotranscriptionally and is stimulated by Pol II CTD ( Proudfoot 2004 ), it is clear that once a particular PAS has been selected and mRNA 3′ cleavage occurs with consequent release from chromatin-associated Pol II, then further cleavage of more proximal PAS on the mRNA will not occur. Thus, mRNAs with extended 3′ UTRs are carried through into the cytoplasm, where particular 3′ UTR sequences act to regulate both the stability and translatability of mRNA as described below (for a recent review on APA, see Lutz and Moreira 2011 ).
What is PAS in eukaryotic genes?
Polyadenylation [poly (A)] signals (PAS) are a defining feature of eukaryotic protein-coding genes. The central sequence motif AAUAAA was identified in the mid-1970s and subsequently shown to require flanking, auxiliary elements for both 3′-end cleavage and polyadenylation of premessenger RNA (pre-mRNA) as well as to promote downstream transcriptional termination. More recent genomic analysis has established the generality of the PAS for eukaryotic mRNA. Evidence for the mechanism of mRNA 3′-end formation is outlined, as is the way this RNA processing reaction communicates with RNA polymerase II to terminate transcription. The widespread phenomenon of alternative poly (A) site usage and how this interrelates with pre-mRNA splicing is then reviewed. This shows that gene expression can be drastically affected by how the message is ended. A central theme of this review is that while genomic analysis provides generality for the importance of PAS selection, detailed mechanistic understanding still requires the direct analysis of specific genes by genetic and biochemical approaches.
When was Escherichia coli DNA polymerase used?
My own research studies as a graduate student at Cambridge, UK, in the mid-1970s with George Brownlee and Fred Sanger began by using Escherichia coli DNA polymerase to partially reverse-transcribe mRNA (retroviral reverse transcriptase had yet to be purified).
Does poly (A) have a nuclear function?
Adesnik M, Salditt M, Thomas W, Darnell JE. 1972. Evidence that all messenger RNA molecules (except histone messenger RNA) contain poly (A) sequences and that the poly (A) has a nuclear function. J Mol Biol 71: 21–30.