What is deletion mapping?
Why is deletion mapping important?
What is the difference between deletion and MAC?
How is deletion mapping carried out?
What are the deletions in the mouse genome?
What is the role of prolactin in transcription?
Can deletions be created in mouse cells?
See 4 more
About this website
What does deletion mean in genetics?
Deletion. A deletion changes the DNA sequence by removing at least one nucleotide in a gene. Small deletions remove one or a few nucleotides within a gene, while larger deletions can remove an entire gene or several neighboring genes. The deleted DNA may alter the function of the affected protein or proteins.
What is the importance of deletion?
Deletions are responsible for an array of genetic disorders, including some cases of male infertility, two thirds of cases of Duchenne muscular dystrophy, and two thirds of cases of cystic fibrosis (those caused by ΔF508). Deletion of part of the short arm of chromosome 5 results in Cri du chat syndrome.
What is deletion and duplication?
Deletions occur when a chromosome breaks and some genetic material is lost. Deletions can be large or small, and can occur anywhere along a chromosome. Duplications. Duplications occur when part of a chromosome is abnormally copied (duplicated).
How do you check for deletions?
Methods commonly used to detect deletion boundaries include long-range PCR and primer walking (Quadri et al., 2015. Fam Cancer 14:41-49.). However, the success of these methods depends on many different factors, including deletion size, GC content and the presence of DNA repeats.
What is deletion with example?
Deletion is the process of erasing or removing something, like the deletion of all the spam emails from your in-box or a newspaper editor's deletion of extraneous information in a draft of an article.
What is another word for deletion?
Synonyms for deletion in English. deletion: erasure; crossing out; cancellation; deletion; removal; striking out; omission; neglect; elision; declaration; oversight; ellipsis; non-attendance; cut; excision.
What is Del DUP analysis?
In clinical genetic diagnostics, deletion/duplication (del/dup) analysis is used to detect disease-causing deletions or duplications in the genome. The size of the alteration may vary from a single exon (<150-1000 bps) to multiple genes, and up to chromosome-level changes visible by light microscopy.
What causes a gene deletion?
Deletions occur when there is homologous but unequal recombination between gene sequences. Similar sequences in the human genome can cross over during mitosis or meiosis, resulting in a shortened portion of the gene sequence.
What is the most common disorder caused by a chromosomal deletion?
1.4. 22q11 deletion syndrome is the most common human chromosomal deletion syndrome occurring in approximately 1 per 4000–6000 live births [32]. Clinical features include learning disabilities/impairments, palate anomalies (including velopharangeal insufficiency (VPI)), characteristic facial appearance (Fig.
Can PCR detect deletions?
The modified PR-PCR method is quite capable of detecting various mutation types, including point mutations and insertions/deletions (indels), and allows discrimination amplification when the mismatch is located within the last eight nucleotides from the 3'-end of the ddNTP-blocked primer.
What disease is caused by deletion mutation?
Deletion. Deletion mutations are actually the cause for a large number of genetic diseases, such as two-thirds of cystic fibrosis cases and the cat cry syndrome, which is so-called because children with this syndrome often have a cry that sounds similar to a cat meowing.
Can Whole Genome Sequencing detect deletions?
Whole-genome sequencing can detect single nucleotide variants, insertions/deletions, copy number changes, and large structural variants.
What is deletion in chromosomal aberration?
What are deletions? The term "deletion" simply means that a part of a chromosome is missing or "deleted." A very small piece of a chromosome can contain many different genes. When genes are missing, there may be errors in the development of a baby since some of the "instructions" are missing.
What causes a deletion mutation?
A deletion mutation occurs when part of a DNA molecule is not copied during DNA replication. This uncopied part can be as small as a single nucleotide or as much as an entire chromosome. The loss of this DNA during replication can lead to a genetic disease.
Is deletion mutation harmful?
Because an insertion or deletion results in a frame-shift that changes the reading of subsequent codons and, therefore, alters the entire amino acid sequence that follows the mutation, insertions and deletions are usually more harmful than a substitution in which only a single amino acid is altered.
What happens when a chromosome is deleted?
Chromosomal deletion syndromes result from loss of parts of chromosomes. They may cause severe congenital anomalies and significant intellectual and physical disability.
Deletion mapping | definition of deletion mapping by Medical dictionary
Definition of deletion mapping in the Medical Dictionary by The Free Dictionary
Spring @DeleteMapping - using @DeleteMapping to map DELETE ... - ZetCode
Spring @DeleteMapping example. The following application uses @DeleteMapping to delete a resource. We use annotations to set up a Spring web application. pom.xml src ├───main │ ├───java │ │ └───com │ │ └───zetcode │ │ ├───config │ │ │ MyWebInitializer.java │ │ │ WebConfig.java │ │ ├───controller ...
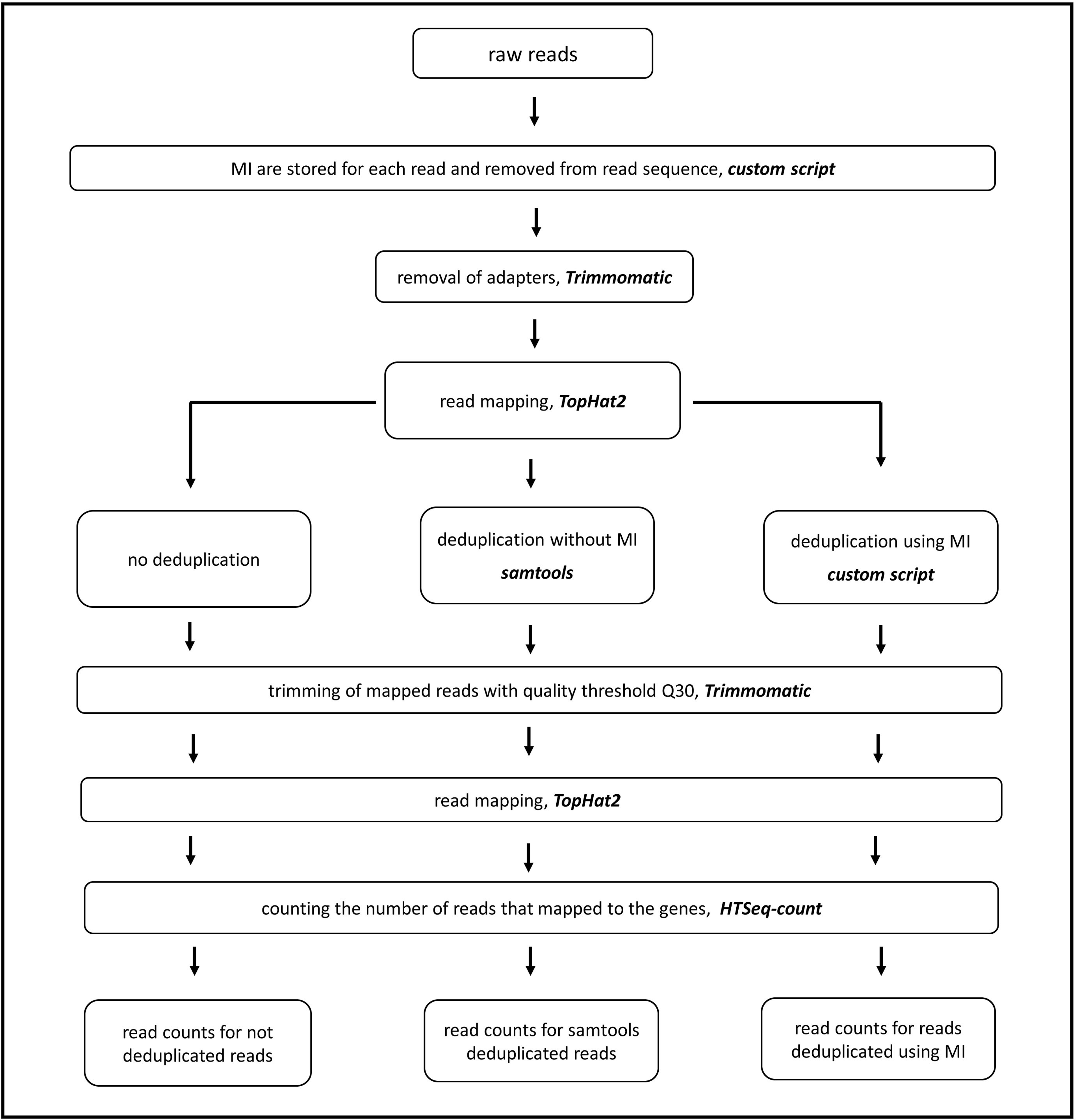
What is deletion mapping?
Deletion mapping is a process by which a DNA feature (mutation, DNA polymorphism, or MIC-limited DNA segment) can be genetically mapped to a segment of a MIC chromosome, defined by deletion ends, using a panel of deletion homo zygotes. These are strains in which both copies of a DNA segment have been deleted from the MIC.
Why is deletion mapping important?
As the above discussions indicate, deletion mapping in the mouse provides a very important complement for fine-structure mapping of genomic regions of <5 cM, where backcross-mapping techniques suffer from low resolution. Deletion mapping is also a preferred method for mapping loci defined by recessive lethal mutations, where it would be difficult to genotype backcross progeny based on phenotype.
What is the difference between deletion and MAC?
To facilitate mapping, most deletion strains are obtained as heterokaryons for drug resistance, generally cycloheximide: they are homozygous in the MIC for the resistance allele, while the MAC is pure for the sensitive allele. True (cross-fertilized) progeny can be easily selected for: the drug resistance derived from the nullisomic strain kills any nonmating parental cells, any cells that mated but retained their parental MAC, and cytogamous progeny derived from the mutant strain, while deletion homozygosity kills cytogamous progeny of the nullisomic strain. It is important that the mutant strain not be expressing the drug resistance encoded in the MIC of the nullisomic strains; otherwise, true progeny cannot be selected. One additional advantage of deletion mapping is that only hemizygous progeny are obtained when the mutation is in the deleted region, so that in mass crosses the informative, mutant progeny cannot be outgrown by wild-type cells before being phenotypically tested, even if homozygosity (or hemizygosity) for the mutation is deleterious and lowers the growth rate.
How is deletion mapping carried out?
Deletion mapping of a recessive mutation is carried out by crossing the homozygous mutant in parallel to every strain of the homozygous deletion panel. For any given deletion cross, F1 progeny show the mutant phenotype only when they are hemizygous for the mutation because it lies within the deleted MIC chromosome segment; otherwise heterozygous progeny with wild-type phenotype are obtained. The locus can then be uniquely assigned to a chromosome arm or smaller deletion interval.
What are the deletions in the mouse genome?
These so-called ‘deletion complexes’ exist for a number of regions of the genome, including the chromosome 7 albino ( c; now called Tyr) and the pink-eyed dilution ( p) loci; the chromosome 4 brown ( b; now called Tyrp1) locus; the chromosome 9 dilute and short-ear loci discussed above; the piebald spotting ( s; now called Ednrb) locus; the chromosome 2 agouti ( a) locus; and several loci mapping within the t region of chromosome 17. At least for the c, p, b, s, and d-se regions, dozens of deletions exist. Genetic analyses of these deletions, incorporating strategies outlined above, as well as several additional strategies described below, have made these chromosomal regions among the functionally best characterized in the mouse genome.
What is the role of prolactin in transcription?
The prolactin gene promoter region is able to confer transcriptional responses to a wide variety of hormones, and deletion mapping has indicated that the responses to, for example, TRH, epidermal growth factor (EGF), dopamine, intracellular calcium, and phorbol ester largely map onto Pit-1-binding elements (e.g. Elsholtz et al., 1986; Day and Maurer, 1989; Yan and Bancroft, 1991; Hoggard et al., 1991 ). However, an additional element adjacent to one of the proximal Pit-1-binding sites is also central to the responses to cyclic AMP and EGF ( Peers et al., 1991; Berwaer et al., 1993 ), and indeed, the estrogen response element must interact with its adjacent Pit-1 element in the distal promoter for full transcriptional effect. Thus, it seems likely that protein–protein interactions involving Pit-1 are important for mediating hormonal regulation. The precise mechanisms for Pit-1 activation of prolactin gene transcription are not yet established, although Pit-1 binding to different DNA elements is modified by phosphorylation by protein kinases A and C ( Kapiloff et al., 1991 ), and Pit-1 gene transcription is regulated by cyclic AMP and by Pit-1 itself ( Chen et al., 1990; McCormick et al., 1990 ).
Can deletions be created in mouse cells?
One major problem in applying the general strategies discussed here efficiently to the entire mouse genome has been that panels of deletions were available only for the specific chromosomal regions outlined above. However, recent encouraging results from several laboratories have indicated that deletion complexes may be created anywhere in the genome by genetic manipulation of embryonic stem (ES) cells. ES cells are derived from the early mouse embryo, can be propagated in tissue culture (and therefore manipulated in vitro ), and, importantly, can be introduced back into developing mouse embryos where they contribute to both somatic and germline tissues. Thus, a mutation introduced into ES cells in vitro can eventually wind up in heterozygotes, and then be bred to homozygosity in a living mouse. Two major strategies have been applied that can induce deletions in ES cells: one involves gene targeting and a Cre-lox protein-mediated intrachromosomal recombination, whereas the other involves exposing ES cells to radiation and selecting, in vitro, for chromosomal deletions. In both cases, mice can eventually be created from ES cells carrying deletions, so that panels of deletions, for fine-structure mapping of the entire genome in the mouse, will be available in the not-so-distant future.
What is the advantage of Listiwse deletion?
The big advantage of listiwse deletion is that it is easy to understand and to implement. In fact, it is the default method in many statistical software such as R or SPSS.
What is listwise deletion?
Definition: Listwise deletion (also known as casewise deletion or complete case analysis) removes all observations from your data, which have a missing value in one or more variables. Complete data without any missing values is needed for many kinds of calculations, e.g. regression or correlation analyses. Listwise deletion is used ...
Which software uses listwise deletion?
Many software packages such as R, SAS, Stata or SPSS use listwise deletion as default method, if nothing else is specified. Even though you might not have heard about listwise or casewise deletion yet, you have probably already used it.
What can be used to replace missings with new values?
More sophisticated methods, e.g. missing data imputation, can be used to replace missings with new values in order to improve data analyses ( Allison, 2002 ).
Where to find the difference compared to an analysis without missing values?
The difference compared to an analysis without missing values can be found in the description of our output: In the description of our regression output in Table 2 you can read the line “ 259 observations deleted due to missingness “.
Can we inspect missing mechanisms?
Be aware that we were able to inspect the missing mechanisms with these statistical tests, since we created the data ourselves. In reality we would not be able to perform such tests and therefore we would need to rely on theoretical assumptions about the randomness of our incomplete data.
Do variables X2 and X3 have missing values?
The variables X2 and X3 do not contain any missing values. This is a typical situation that you may also find in reality. Missing rates of 20 or more percent are nothing special in voluntary surveys. Especially sensitive questions about income, sexual orientation, or age are at risk for high nonresponse rates.
What is deletion mapping?
Deletion mapping is a process by which a DNA feature (mutation, DNA polymorphism, or MIC-limited DNA segment) can be genetically mapped to a segment of a MIC chromosome, defined by deletion ends, using a panel of deletion homo zygotes. These are strains in which both copies of a DNA segment have been deleted from the MIC.
Why is deletion mapping important?
As the above discussions indicate, deletion mapping in the mouse provides a very important complement for fine-structure mapping of genomic regions of <5 cM, where backcross-mapping techniques suffer from low resolution. Deletion mapping is also a preferred method for mapping loci defined by recessive lethal mutations, where it would be difficult to genotype backcross progeny based on phenotype.
What is the difference between deletion and MAC?
To facilitate mapping, most deletion strains are obtained as heterokaryons for drug resistance, generally cycloheximide: they are homozygous in the MIC for the resistance allele, while the MAC is pure for the sensitive allele. True (cross-fertilized) progeny can be easily selected for: the drug resistance derived from the nullisomic strain kills any nonmating parental cells, any cells that mated but retained their parental MAC, and cytogamous progeny derived from the mutant strain, while deletion homozygosity kills cytogamous progeny of the nullisomic strain. It is important that the mutant strain not be expressing the drug resistance encoded in the MIC of the nullisomic strains; otherwise, true progeny cannot be selected. One additional advantage of deletion mapping is that only hemizygous progeny are obtained when the mutation is in the deleted region, so that in mass crosses the informative, mutant progeny cannot be outgrown by wild-type cells before being phenotypically tested, even if homozygosity (or hemizygosity) for the mutation is deleterious and lowers the growth rate.
How is deletion mapping carried out?
Deletion mapping of a recessive mutation is carried out by crossing the homozygous mutant in parallel to every strain of the homozygous deletion panel. For any given deletion cross, F1 progeny show the mutant phenotype only when they are hemizygous for the mutation because it lies within the deleted MIC chromosome segment; otherwise heterozygous progeny with wild-type phenotype are obtained. The locus can then be uniquely assigned to a chromosome arm or smaller deletion interval.
What are the deletions in the mouse genome?
These so-called ‘deletion complexes’ exist for a number of regions of the genome, including the chromosome 7 albino ( c; now called Tyr) and the pink-eyed dilution ( p) loci; the chromosome 4 brown ( b; now called Tyrp1) locus; the chromosome 9 dilute and short-ear loci discussed above; the piebald spotting ( s; now called Ednrb) locus; the chromosome 2 agouti ( a) locus; and several loci mapping within the t region of chromosome 17. At least for the c, p, b, s, and d-se regions, dozens of deletions exist. Genetic analyses of these deletions, incorporating strategies outlined above, as well as several additional strategies described below, have made these chromosomal regions among the functionally best characterized in the mouse genome.
What is the role of prolactin in transcription?
The prolactin gene promoter region is able to confer transcriptional responses to a wide variety of hormones, and deletion mapping has indicated that the responses to, for example, TRH, epidermal growth factor (EGF), dopamine, intracellular calcium, and phorbol ester largely map onto Pit-1-binding elements (e.g. Elsholtz et al., 1986; Day and Maurer, 1989; Yan and Bancroft, 1991; Hoggard et al., 1991 ). However, an additional element adjacent to one of the proximal Pit-1-binding sites is also central to the responses to cyclic AMP and EGF ( Peers et al., 1991; Berwaer et al., 1993 ), and indeed, the estrogen response element must interact with its adjacent Pit-1 element in the distal promoter for full transcriptional effect. Thus, it seems likely that protein–protein interactions involving Pit-1 are important for mediating hormonal regulation. The precise mechanisms for Pit-1 activation of prolactin gene transcription are not yet established, although Pit-1 binding to different DNA elements is modified by phosphorylation by protein kinases A and C ( Kapiloff et al., 1991 ), and Pit-1 gene transcription is regulated by cyclic AMP and by Pit-1 itself ( Chen et al., 1990; McCormick et al., 1990 ).
Can deletions be created in mouse cells?
One major problem in applying the general strategies discussed here efficiently to the entire mouse genome has been that panels of deletions were available only for the specific chromosomal regions outlined above. However, recent encouraging results from several laboratories have indicated that deletion complexes may be created anywhere in the genome by genetic manipulation of embryonic stem (ES) cells. ES cells are derived from the early mouse embryo, can be propagated in tissue culture (and therefore manipulated in vitro ), and, importantly, can be introduced back into developing mouse embryos where they contribute to both somatic and germline tissues. Thus, a mutation introduced into ES cells in vitro can eventually wind up in heterozygotes, and then be bred to homozygosity in a living mouse. Two major strategies have been applied that can induce deletions in ES cells: one involves gene targeting and a Cre-lox protein-mediated intrachromosomal recombination, whereas the other involves exposing ES cells to radiation and selecting, in vitro, for chromosomal deletions. In both cases, mice can eventually be created from ES cells carrying deletions, so that panels of deletions, for fine-structure mapping of the entire genome in the mouse, will be available in the not-so-distant future.
