
What is the KD of an enzyme?
The Kd of an enzymatic reaction expresses the ligand-receptor affinity. In other words, it states the capability of a substrate to leave the receptor of an enzyme. On the other hand, it describes how strongly a substrate binds to the enzyme. What is Km? Km is the Michaelis constant. Unlike Kd, Km is a kinetic constant.
What is the difference between KD and km in chemistry?
Kd and Km are equilibrium constants. The key difference between Kd and Km is that Kd is a thermodynamic constant whereas Km is not a thermodynamic constant. Kd refers to dissociation constant while Km is the Michaelis constant. Both these constants are very important in the quantitative analysis of enzymatic reactions.
What is the reaction rate constant (Kd)?
The measurement of the reaction rate constants can be used to define an equilibrium or affinity constant (1/K D ). In short, the smaller the K D value the greater the affinity of the antibody for its target.
What is meant by km in enzyme kinetics?
Thus, KM is the substrate concentration at which the reaction velocity is half of the maximum velocity. The two important properties of enzyme kinetics is how easily can the enzyme be saturated with a substrate, and the maximum rate it can achieve.
What does Kd mean in enzyme kinetics?
dissociation constantThe dissociation constant (Kd) The dissociation constant (Kd) quantifies the equilibrium between a ligand (L) being free in solution and bound to a site in a protein (EL): It corresponds to the affinity which the ligand has for the binding site.
What does Kd value mean?
The KD value relates to the concentration of antibody (the amount of antibody needed for a particular experiment) and so the lower the KD value (lower concentration) and thus the higher the affinity of the antibody. KD value. Molar concentration (sensitivity) 10-4 to 10-6. Micromolar (µM)
Why is Kd important?
So a higher Kd means that when you go take a molecular census, there are more unbound molecules, whereas a lower Kd means that you find more bound molecules.
What is Kd in chemistry?
Dissociation constant (KD) defined as amount of reactant that dissociates reversibly to form component products. From: Encyclopedia of Biological Chemistry (Third Edition), 2021.
What is enzyme kinetics?
Enzyme kinetics is the branch of biochemistry that deals with a quantitative description of this process, mainly, how experimental variables affect reaction rates. The variables that are studied include the concentrations of the enzymes, substrates (reactants), products, inhibitors, activators, the pH, temperature, and ionic strength.
When was enzyme kinetics developed?
Enzyme kinetic principles, developed early in the twentieth century, were based on homogeneous systems, this is, when the biocatalyst and their substrates and products of reaction are in a single phase where the reaction occurs.
What is the purpose of enzyme crystals?
Enzyme crystals used for resolving three-dimensional (3D) structures (via X-ray diffraction or nuclear magnetic resonance (NMR)) need amounts of purified enzymes that frequently are difficult to isolate from insects by conventional separation procedures.
What is kinetic analysis?
A complete kinetic analysis (together with complementary studies of equilibrium ligand binding, isotope exchange, covalent modification of amino acid side chains, etc.) can disclose most of the functional characteristics of a particular enzym e. These include (1) the specificities and affinities of the ligand subsites, ...
Can cDNA be used for mutagenesis?
Alternatively, cDNA may be amenable to site-directed mutagenesis for structure/function studies. Site-directed mutagenesis tests the role of individual amino acid residues in enzyme function or structure. Such knowledge is a prerequisite in developing new biotechnological approaches to control insects via the gut.
What is enzyme kinetics?
Enzyme kinetics is the study of the rates of enzyme-catalysed chemical reactions. In enzyme kinetics, the reaction rate is measured and the effects of varying the conditions of the reaction are investigated. Studying an enzyme's kinetics in this way can reveal the catalytic mechanism of this enzyme, its role in metabolism, ...
Who developed the kinetics of enzymes?
Maud Leonora Menten (a postdoctoral researcher in Michaelis's lab at the time) repeated Henri's experiments and confirmed his equation, which is now generally referred to as Michaelis-Menten kinetics (sometimes also Henri-Michaelis-Menten kinetics ). Their work was further developed by G. E. Briggs and J. B. S. Haldane, who derived kinetic equations that are still widely considered today a starting point in modeling enzymatic activity.
Why is kinetics important?
Firstly, it helps explain how enzymes work, and secondly, it helps predict how enzymes behave in living organisms. The kinetic constants defined above, KM and Vmax, are critical to attempts to understand how enzymes work together to control metabolism .
How do enzyme inhibitors inactivate enzymes?
Enzyme inhibitors can also irreversibly inactivate enzymes, usually by covalently modifying active site residues. These reactions, which may be called suicide substrates, follow exponential decay functions and are usually saturable. Below saturation, they follow first order kinetics with respect to inhibitor. Irreversible inhibition could be classified into two distinct types. Affinity labelling is a type of irreversible inhibition where a functional group that is highly reactive modifies a catalytically critical residue on the protein of interest to bring about inhibition. Mechanism-based inhibition, on the other hand, involves binding of the inhibitor followed by enzyme mediated alterations that transform the latter into a reactive group that irreversibly modifies the enzyme.
What enzymes produce oxaloacetate?
For example, oxaloacetate is formed by malate dehydrogenase within the mitochondrion. Oxaloacetate can then be consumed by citrate synthase, phosphoenolpyruvate carboxykinase or aspartate aminotransferase, feeding into the citric acid cycle, gluconeogenesis or aspartic acid biosynthesis, respectively.
What is enzyme assay?
Enzyme assays are laboratory procedures that measure the rate of enzyme reactions. Since enzymes are not consumed by the reactions they catalyse, enzyme assays usually follow changes in the concentration of either substrates or products to measure the rate of reaction. There are many methods of measurement.
What is an enzyme?
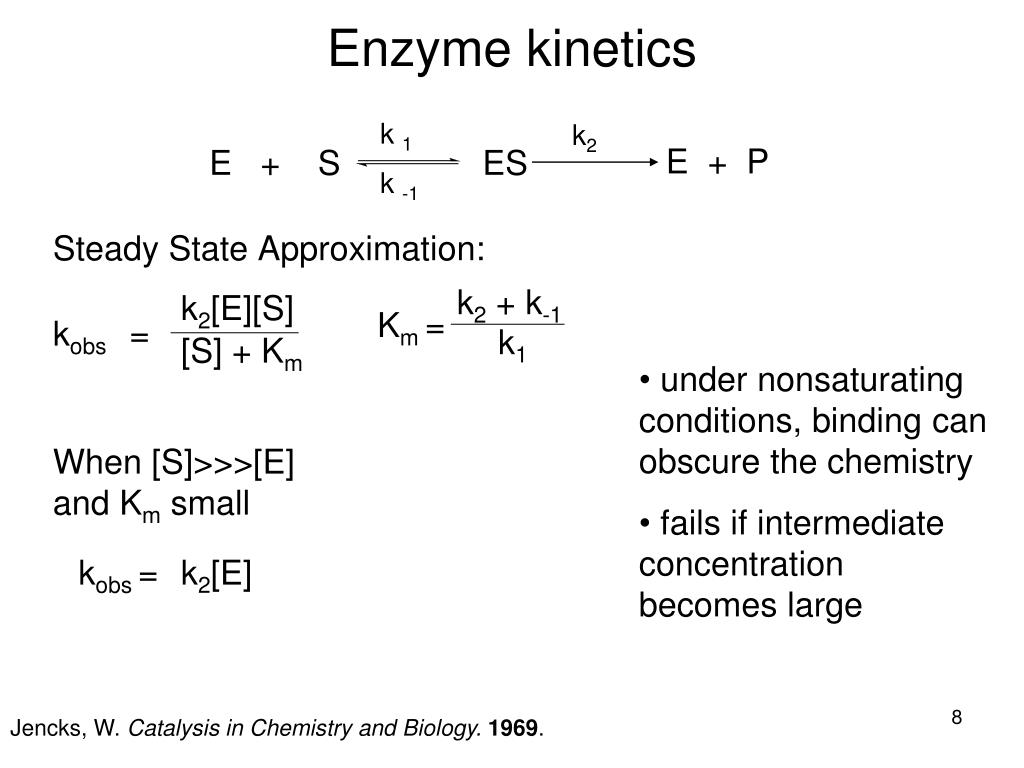
An enzyme (E) is typically a protein molecule that promotes a reaction of another molecule, its substrate (S). This binds to the active site of the enzyme to produce an enzyme-substrate complex ES, and is transformed into an enzyme-product complex EP and from there to product P, via a transition state ES*.
What is KD in chemistry?
What is Kd? Kd is dissociation constant. It is also known as equilibrium dissociation constant due to its use in equilibrium systems. The dissociation constant is the equilibrium constant of reactions where a large compound is converted into small components reversibly.
What is the difference between KD and KM?
The key difference between Kd and Km is that Kd is a thermodynamic constant whereas Km is not a thermodynamic constant. Kd refers to dissociation constant while Km is the Michaelis constant. Both these constants are very important in the quantitative analysis of enzymatic reactions.
What is the equation of dissociation constant?
The equation of dissociation constant, Kd for the above reaction is as follows: Specifically, in biochemical applications, Kd helps to determine the amount of products given by a chemical reaction in the presence of an enzyme. The Kd of an enzymatic reaction expresses the ligand-receptor affinity.
What is the difference between KD and KD?
Ki refers to inhibition constant, while Kd means dissociation constant. Both terms are used to describe the binding affinity that a small molecule or macromolecule has for an enzyme or receptor. The difference is that Kd is a more general, all-encompassing term. As discussed in my article on the difference between Km and Kd, ...
What is the Ki constant?
The Ki inhibition constant also represents a dissociation constant, but more narrowly for the binding of an inhibitor to an enzyme. That is, a ligand whose binding reduces the catalytic activity of the enzyme. The binding equilibrium described by the Ki value depends on the kinetic mechanism of inhibition.
Why is IC50 less precise than Ki?
Because it does not directly measure a binding equilibrium, IC50 is less precise than Ki or Kd. Importantly, the values obtained are highly dependent on the measurement conditions and the mechanism of inhibition. The simplicity and lack of assumptions required to generate these data make them useful.
What is K D and how does it correlate to antibody affinity and sensitivity?
K D is the equilibrium dissociation constant, a ratio of k off /k on, between the antibody and its antigen. K D and affinity are inversely related.
How were K D values measured?
The measurements were undertaken as a collaboration with J. Landry and X. Zhu, Dept. of Physics, UC Davis. Data processing, data analysis and report generation for OI-RD (Oblique-incidence reflectivity difference) binding affinity data were measured at UC Davis, Dept.
How to read K D value
The specific K D values and individual plots for each antibody have been added to each individual antibody product page for you to be able to see each antibodies measured K D value.
What does the K D value represent? How is it calculated?
K D is the ratio of the antibody dissociation rate (k off ), how quickly it dissociates from its antigen, to the antibody association rate (k on) of the antibody, how quickly it binds to its antigen.
What is the relationship between K D and antibody affinity?
Affinity is the strength of binding of a single molecule to its ligand. It is typically measured and reported by the equilibrium dissociation constant (K D ), which is used to evaluate and rank order strengths of bimolecular interactions.
What is the typical K D value for an antibody? What would one expect to be a good K D value?
Most antibodies have K D values in the low micromolar (10 -6) to nanomolar (10 -7 to 10 -9) range. High affinity antibodies generally considered to be in the low nanomolar range (10 -9) with very high affinity antibodies being in the picomolar (10 -12) range.
How were the K D values measured?
The K D values were measured using a novel label-free detection system developed by Dr. James Landry and Dr. Xiangdong Zhu at the University of California Davis, Department of Physics.

Overview
Single-substrate reactions
Enzymes with single-substrate mechanisms include isomerases such as triosephosphateisomerase or bisphosphoglycerate mutase, intramolecular lyases such as adenylate cyclase and the hammerhead ribozyme, an RNA lyase. However, some enzymes that only have a single substrate do not fall into this category of mechanisms. Catalase is an example of this, as the enzyme reacts with a first molecule of hydrogen peroxide substrate, becomes oxidised and is the…
General principles
The reaction catalysed by an enzyme uses exactly the same reactants and produces exactly the same products as the uncatalysed reaction. Like other catalysts, enzymes do not alter the position of equilibrium between substrates and products. However, unlike uncatalysed chemical reactions, enzyme-catalysed reactions display saturation kinetics. For a given enzyme concentration and for relatively low substrate concentrations, the reaction rate increases linearly with substrate conce…
Enzyme assays
Enzyme assays are laboratory procedures that measure the rate of enzyme reactions. Since enzymes are not consumed by the reactions they catalyse, enzyme assays usually follow changes in the concentration of either substrates or products to measure the rate of reaction. There are many methods of measurement. Spectrophotometric assays observe change in the absorbance of light between products and reactants; radiometric assays involve the incorporation or release of
Multi-substrate reactions
Multi-substrate reactions follow complex rate equations that describe how the substrates bind and in what sequence. The analysis of these reactions is much simpler if the concentration of substrate A is kept constant and substrate B varied. Under these conditions, the enzyme behaves just like a single-substrate enzyme and a plot of v by [S] gives apparent KM and Vmax constants for substrate B. If a set of these measurements is performed at different fixed concentrations o…
Reversible catalysis and the Haldane equation
External factors may limit the ability of an enzyme to catalyse a reaction in both directions (whereas the nature of a catalyst in itself means that it cannot catalyse just one direction, according to the principle of microscopic reversibility). We consider the case of an enzyme that catalyses the reaction in both directions:
The steady-state, initial rate of the reaction is
Non-Michaelis–Menten kinetics
Many different enzyme systems follow non Michaelis-Menten behavior. A select few examples include kinetics of self-catalytic enzymes, cooperative and allosteric enzymes, interfacial and intracellular enzymes, processive enzymes and so forth. Some enzymes produce a sigmoid v by [S] plot, which often indicates cooperative binding of substrate to the active site. This means that the binding of one substrate molecule affects the binding of subsequent substrate molecules. This …
Pre-steady-state kinetics
In the first moment after an enzyme is mixed with substrate, no product has been formed and no intermediates exist. The study of the next few milliseconds of the reaction is called pre-steady-state kinetics. Pre-steady-state kinetics is therefore concerned with the formation and consumption of enzyme–substrate intermediates (such as ES or E*) until their steady-state concentrations are reached.