
What is the neighbor-joining method?
Apr 02, 2022 · The neighbor-joining method is a special case of The neighbor-joining method: a new method for reconstructing phylogenetic trees. The principle of this method is to find pairs of operational taxonomic units (OTUs [= neighbors]) that minimize the total branch length at each stage of clustering of OTUs starting with a starlike tree.
What is neighbor joining tree?
The neighbor-joining method: a new method for reconstructing phylogenetic trees. A new method called the neighbor-joining method is proposed for reconstructing phylogenetic trees from evolutionary distance data. The principle of this method is to find pairs of operational taxonomic units (OTUs [= neighbors]) that minimize the total branch length at each stage of clustering of …
What is neighbor joining in biology?
The neighbor-joining method: a new method for reconstructing phylogenetic trees. The principle of this method is to find pairs of operational taxonomic units (OTUs [= neighbors]) that minimize the total branch length at each stage of clustering of OTUs starting with a starlike tree.
What are the properties of neighbor joining?
The neighbor-joining method is a special case of the star decomposition method. In contrast to cluster analysis neighbor-joining keeps track of nodes on a tree rather than taxa or clusters of taxa. The raw data are provided as a distance matrix and the initial tree is a star tree. Then a modified distance matrix is constructed in which the separation between each pair of nodes is …

What is the neighbor-joining method?
In bioinformatics, neighbor joining is a bottom-up (agglomerative) clustering method for the creation of phylogenetic trees, created by Naruya Saitou and Masatoshi Nei in 1987.
What is neighbor-joining method of phylogenetic tree construction?
Neighbor-Joining (NJ) tree inference method was originally written by Saitou and Nei in 1987. It belongs to a class of distance-based methods used to build evolutionary trees. NJ method takes a matrix of pairwise evolutionary distances between the given sequences to build the evolutionary tree.Dec 27, 2017
Is neighbor-joining distance based?
The neighbor-joining method (NJ) is a distance based method (requires a distance matrix) and uses the star decomposition method. Neighbor-joining is a recursive algorithm. Each step in the recursion consists of the following steps: 1.
What is the difference between UPGMA and neighbor-joining?
The key difference between UPGMA and neighbor joining tree is the type of the phylogenetic tree resulting from each method. UPGMA is the technique of constructing a rooted phylogenetic tree while neighbor joining tree is the technique of constructing an unrooted phylogenetic tree.Oct 15, 2019
How do I make my Neighbour join a tree?
0:122:35Making a Neighbor-Joining Tree with MEGA5 (Part 1) - YouTubeYouTubeStart of suggested clipEnd of suggested clipSeveral common sequence data file formats. So this process works well to import your data into megaMoreSeveral common sequence data file formats. So this process works well to import your data into mega go to the file menu in the mecca window. And choose convert file format to mega.
Why is neighbor joining not the most robust of ways to infer phylogenetic relations?
Neighbour Joining is not a phylogenetic method, but a phenetic one. It establish relationships between sequences according to their genetic distance (a phenetic criteria) alone, without taking into account an evolutionary model. Thus, ancestry is never considered.Oct 30, 2014
When can I use Neighbour joining?
1:4622:59Phylogenetics Part 4 - Neighbor Joining Method - YouTubeYouTubeStart of suggested clipEnd of suggested clipLet's try to understand neighbor-joining method using using an example here we have four differentMoreLet's try to understand neighbor-joining method using using an example here we have four different sequences a b c and d and all sequences work compared to each other.
Is a method of Reconstructingevolutionary trees?
A new method called the neighbor-joining method is proposed for reconstructing phylogenetic trees from evolutionary distance data.
What is UPGMA in bioinformatics?
UPGMA: Unweighted Pair Group Method with Arithmetic Mean: A simple clustering method that assumes a constant rate of evolution (molecular clock hypothesis).
What is Ultrametric distance?
Ultrametrics are a special case of tree metrics. A distance function on a finite set is a tree metric if there exists a weighted tree, whose vertex set contains , such that the distance between any two points in is equal to the length of the unique path connecting them in the tree.Jan 10, 2015
Why is UPGMA an unreliable method?
UPGMA is the simplest method for constructing trees. The great disadvantage of UPGMA is that it assumes the same evolutionary speed on all lineages, i.e. the rate of mutations is constant over time and for all lineages in the tree. This is called a 'molecular clock hypothesis'.
Is a UPGMA tree rooted or unrooted?
The most important practical issues: UPGMA provides rooted tree as a result, while NJ unrooted, and you have to take care proper rooting the NJ tree afterward.Mar 17, 2014
What is neighbor joining?
In bioinformatics, neighbor joining is a bottom-up (agglomerative) clustering method for the creation of phylogenetic trees , created by Naruya Saitou and Masatoshi Nei in 1987. Usually used for trees based on DNA or protein sequence data, the algorithm requires knowledge of the distance between each pair of taxa (e.g., species or sequences) to form the tree.
Is NJ fast?
The main virtue of NJ is that it is fast:466 as compared to least squares, maximum parsimony and maximum likelihood methods. This makes it practical for analyzing large data sets (hundreds or thousands of taxa) and for bootstrapping, for which purposes other means of analysis (e.g. maximum parsimony, maximum likelihood) may be computationally prohibitive.
Is a distance matrix additive?
A distance matrix whose distances agree in this way with some tree is said to be 'additive', a property which is rare in practice. Nonetheless it is important to note that, given an additive distance matrix as input, neighbor joining is guaranteed to find the tree whose distances between taxa agree with it.
What is neighbor-joining tree?
On the other hand, the neighbor-joining tree is the second method used to build a phylogenetic tree from a distance matrix. However, it produces an unrooted phylogenetic tree whose branch lengths reflect the amount of change during the evolution.
What is the difference between neighbor joining tree and UPGMA?
The main difference between UPGMA and neighbor joining tree is that UPGMA is an agglomerative hierarchical clustering method based on the average linkage method whereas neighbor-joining tree is an iterative clustering method based on the minimum-evolution criterion. Furthermore, UPGMA produces a rooted phylogenetic tree while neighbor-jo ining tree method produces an unrooted phylogenetic tree. Since UPGMA method assumes equal rates of evolution, branch tips come out equal while as neighbor-joining tree method allows unequal rates of evolution, the branch lengths are proportional to the amount of change.
What is UPGMA algorithm?
UPGMA is one of the two algorithms to build a phylogenetic tree based on the evolutionary distance data. Moreover, it builds a rooted phylogenetic tree with similar branch lengths. In addition, it is the simple, fast, and the most reliable algorithm for building a phylogenetic tree from distance matrices. On the other hand, the neighbor-joining ...
What is the nature of branches in a phylogenetic tree?
Nature of Branches of the Phylogenetic Tree. As UPGMA method assumes equal rates of evolution, branch tips come out equal (same branch length from the root to the tips). As neighbor-joining tree method allows unequal rates of evolution, the branch lengths are proportional to the amount of change.
What is UPGMA in physics?
What is UPGMA. UPGMA (unweighted pair group method with arithmetic mean) is a simple, agglomerative, hierarchical clustering method attributed to Sokal and Michener. It is the simplest and fastest method for building a rooted and ultrametric phylogenetic tree.
What is Hasa's degree?
Hasa has a BA degree in English, French and Translation studies. She is currently reading for a Masters degree in English. Her areas of interests include literature, language, linguistics and also food.
What is the major drawback of the Molecular Clock Hypothesis?
However, the major drawback of the method is its assumption of the same evolutionary rate on all lineages. This means the rate of mutations in these lineages is constant over time. This is also called the ‘molecular clock hypothesis’. In addition, it produces all the branches in the tree with similar distances.
What does NJ mean in phylogenetics?
NJ assumes that species with lower difference in the given matrix must be closer than those species with any other on the matrix. You can use NJ as your phylogenetic method, but usually only matrices with low distance between sequences have good results with NJ. MEGA can provide you with p distances for each species and if ...
Which is better, NJ or ML?
NJ is faster in term of computing but quite weaker compare to ML... ML uses more complex evolution model, so "closer to the real life" and the mathematics behind ML/bayesian methods are know to be stronger than NJ for reconstructing sequence histories.
What is genetic distance?
Genetic distances are then considered as estimates of the number of mutations that have accumulated along the branches leading to each sequence since they split from their common ancestor . In ML, the same substitution models are used to calculate the likelihood of a specific phylogenetic tree, given the sequence data.
Can Mega be used for phylogeny?
Or only as a pedagogic figure.... Yes, MEGA can be used for publishing phylogeny. Neighbour Joining is not a phylogenetic method, but a phenetic one.
Is neighbour joining phylogenetic or phenetic?
Université de Montpellier. Neighbour Joining is not a phylogenetic method, but a phenetic one. It establish relationships between sequences according to their genetic distance (a phenetic criteria) alone, without taking into account an evolutionary model. Thus, ancestry is never considered.
What is the difference between neighbor joining tree and UPGMA?
The key difference between UPGMA and neighbor joining tree is the type of the phylogenetic tree resulting from each method. UPGMA is the technique of constructing a rooted phylogenetic tree while neighbor joining tree is the technique of constructing an unrooted phylogenetic tree.
What is Dr. Samanthi Udayangani's degree?
Dr.Samanthi Udayangani holds a B.Sc. Degree in Plant Science, M.Sc. in Molecular and Applied Microbiology, and PhD in Applied Microbiology. Her research interests include Bio-fertilizers, Plant-Microbe Interactions, Molecular Microbiology, Soil Fungi, and Fungal Ecology.
What is a phylogenetic tree?
Phylogenetic trees are tree-like diagrams that show evolutionary relationships between organisms. A phylogenetic tree can have different topologies depending on the technique used for tree construction. UPGMA and neighbor joining tree are two main methods that build phylogenetic trees.
Who developed the phylogenetic tree?
It is a hierarchical grouping method. The method was introduced by Sokal and Michener. It is the fastest technique that develops a phylogenetic tree. The resulting phylogenetic tree is a rooted phylogenetic tree with a common ancestor.
Is the UPGMA method accurate?
Therefore, in the real context, the phylogenetic tree constructed from a UPGMA method may not be accurate and reliable. UPGMA method considers pair-wise distances to produce a phylogenetic tree. Initially, each species is a cluster, and two such clusters with the smallest evolutionary distance form a pair.
What is neighbor joining?
Neighbor joining is just a clustering algorithm that clusters haplotypes based on genetic distance and is not often used for publication in recent literature. "Neighbor joining and UPGMA are clustering algorithms that can make quick trees but are not the most reliable, especially when dealing with deeper divergence times.
Which method is used to build a phylogeny using sequence data?
Maximum likelihood and Bayesian methods can apply a model of sequence evolution and are ideal for building a phylogeny using sequence data. These methods are the two methods that are most often used in publications and many reviewers prefer these methods.
Why do people do more than ML?
The main reason people do more than ML is that a lot of elderly (male) reviewers are still adhering to the Holy Parsimony Church of Cladistics or feel more confident seeing all done because they themselves are not aware why different optimality criteria can get different results. In my work ( https://scholar.google.com/citations?
Overview
In bioinformatics, neighbor joining is a bottom-up (agglomerative) clustering method for the creation of phylogenetic trees, created by Naruya Saitou and Masatoshi Nei in 1987. Usually used for trees based on DNA or protein sequence data, the algorithm requires knowledge of the distance between each pair of taxa (e.g., species or sequences) to form the tree.
The algorithm
Neighbor joining takes as input a distance matrix specifying the distance between each pair of taxa. The algorithm starts with a completely unresolved tree, whose topology corresponds to that of a star network, and iterates over the following steps until the tree is completely resolved and all branch lengths are known:
Example
Let us assume that we have five taxa and the following distance matrix :
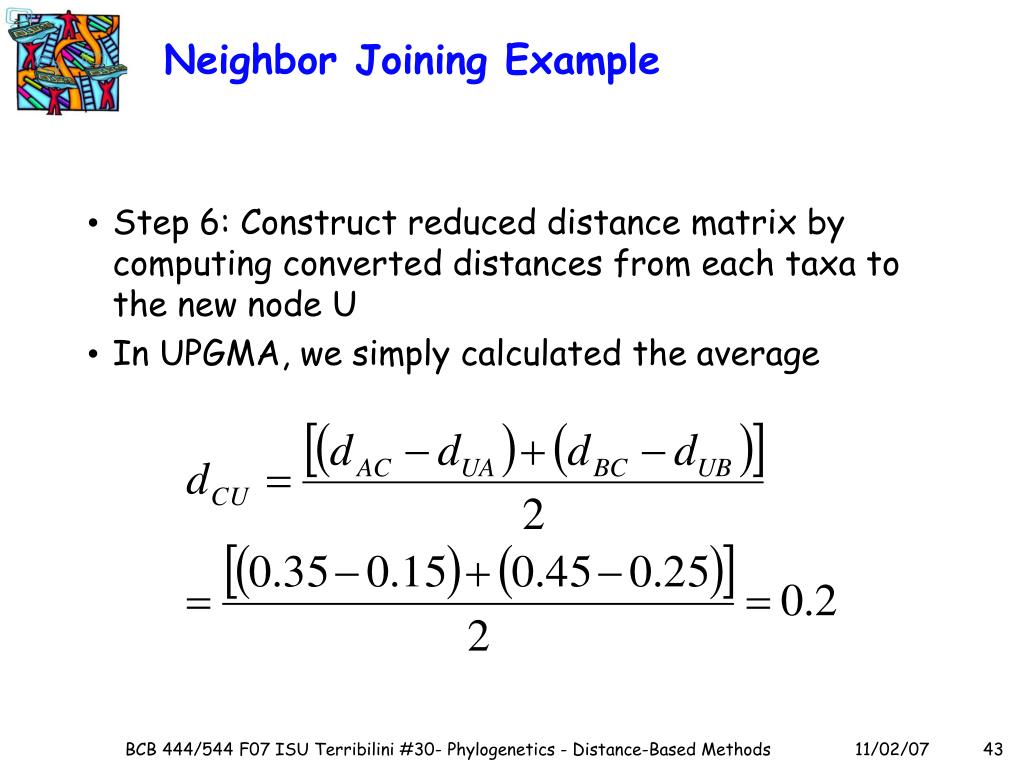
We calculate the values by equation (1). For example:
We obtain the following values for the matrix (the diagonal elements of the matrix are not used and are omitted here):
In the example above, . This is the smallest value of , so we join elements and .
Neighbor joining as minimum evolution
Neighbor joining may be viewed as a greedy heuristic for the Balanced Minimum Evolution(BME) criterion. For each topology, BME defines the tree length (sum of branch lengths) to be a particular weighted sum of the distances in the distance matrix, with the weights depending on the topology. The BME optimal topology is the one which minimizes this tree length. Neighbor joining at each step greedily joins that pair of taxa which will give the greatest decrease in the es…
Advantages and disadvantages
The main virtue of NJ is that it is fast as compared to least squares, maximum parsimony and maximum likelihood methods. This makes it practical for analyzing large data sets (hundreds or thousands of taxa) and for bootstrapping, for which purposes other means of analysis (e.g. maximum parsimony, maximum likelihood) may be computationally prohibitive.
Neighbor joining has the property that if the input distance matrix is correct, then the output tree …
Implementations and variants
There are many programs available implementing neighbor joining. RapidNJ and NINJA are fast implementations with typical run times proportional to approximately the square of the number of taxa. BIONJ and Weighbor are variants of neighbor joining which improve on its accuracy by making use of the fact that the shorter distances in the distance matrix are generally better known than the longer distances. FastMEis an implementation of the closely related balanced minimum evol…
See also
• Nearest neighbor search
• UPGMA and WPGMA
• Minimum Evolution
External links
• The Neighbor-Joining Method — a tutorial