What is the catalytic triad of a serine protease?
The triad is located in the active site of the enzyme, where catalysis occurs, and is preserved in all superfamilies of serine protease enzymes. The triad is a coordinated structure consisting of three amino acids: His 57, Ser 195 (hence the name "serine protease") and Asp 102.
What is a catalytic triad of chymotrypsin?
A catalytic triad is a group of three amino acids that are found in the active sites of some proteases involved in catalysis. Three different proteases that have catalytic triads are: chymotrypsin, trypsin and elastase. In chymotrypsin, the catalytic triad is made from serine 195, histidine 57, and aspartate 102.
Is chymotrypsin a serine protease?
Chymotrypsin: >Used as an example of a serine protease because it's structure and mechanism are well understood. > Catalyzes the hydrolysis of peptide bonds, on the carboxyl side of bulky aromatic side chains (Tyr, Phe, Trp).
Which three amino acids form the catalytic triad of the serine protease chymotrypsin?
Chymotrypsin contains a collection of three amino acids called the catalytic triad. This triad consists of serine-195, histidine-57 and aspartate-102. These amino acids work together to carry out the catalytic function of breaking peptide bonds.
What type of enzyme is chymotrypsin?
It uses an active serine residue to perform hydrolysis on the C-terminus of the aromatic amino acids of other proteins. Chymotrypsin is a protease enzyme that cleaves on the C-terminal phenylalanine (F), tryptophan (W), and tyrosine (Y) on peptide chains.
What is a catalytic triad and how does it work?
Catalytic triads perform covalent catalysis using a residue as a nucleophile. The reactivity of the nucleophilic residue is increased by the functional groups of the other triad members. The nucleophile is polarised and oriented by the base, which is itself bound and stabilised by the acid.
What are examples of serine proteases?
Some examples of serine proteases are:Chymotrypsin - pancreatic digestive enzyme.Trypsin - pancreatic digestive enzyme.Elastase - pancreatic digestive enzyme.Plasmin - dissolves blood clots.Thrombin - activates fibrinogen to form blood clots.Acrosomal protease - sperm penetration of ova.More items...
Why are they called serine proteases?
SERINE PROTEINASES They are so called because they have a catalytically essential serine residue at their active sites. Serine proteinases are optimally active at neutral pH and play major roles in extracellular proteolysis.
Which factors are serine proteases?
Factor X, also known as Stuart-Prower factor, is a serine protease of the coagulation cascade. In the presence of calcium and phospholipid, FⅩ functions in both intrinsic and extrinsic pathway of blood coagulation. FⅩ is activated to FⅩa by factors FIX and F VII.
How many serine proteases are there?
The serine proteases are divided into two families: the trypsins and the subtilisins. The trypsin family is the largest and contains, among others, trypsin and chymotrypsin, elastase, mast cell tryptase, and many of the factors regulating blood coagulation and fibrinolysis.
What three amino acids are found in the catalytic triad of chymotrypsin quizlet?
Serine 195, histidine 57, and aspartate 192 function in the catalytic triad of chymotrypsin. Hydrogen atoms are passed back and forth between the three amino acids.
Why are serine proteases important?
These are digestive enzymes capable of cutting peptide bonds in a wide range of proteins. In some pathways, such as blood clotting or the immune system, a serine protease may be so specific that it only can cut a single peptide bond in a single unique protein substrate.
What is the purpose of the chymotrypsin?
Chymotrypsin is a digestive proteolytic enzyme produced by the pancreas that is used in the small intestine to help digest proteins. The enzyme is also used to help create medicines and has been used in clinical healthcare settings since the 1960s.
What is the role of histidine in the catalytic triad?
Histidine in the catalytic triad has a double role: making oxygen of serine more nucleophilic to attack the carbonyl of the peptide bond and then, in a second step, making water molecule more nucleophilic to attack carbonyl of the acyl enzyme.
What peptide bonds does chymotrypsin cleave?
Chymotrypsin (EC 3.4. 21.1) is a 26kDa serine carboxypeptidase that preferentially cleaves the amide bond (the P1 position) of an aromatic amino acid residues such as tyrosine, tryptophan and phenylalanine.
What determines the specificity of chymotrypsin?
The Active Site Environment A specific pocket adjacent to the active site triad determines the specificity of the protease (chymotrypsin cleaves adjacent to large aromatic side chains, trypsin adjacent to lys or arg residues).
What is the catalytic triad of residues in TEV protease?
The enzyme TEV protease contains an example of a catalytic triad of residues (red) in its active site. The triad consists of an aspartate ( acid ), histidine ( base) and serine ( nucleophile ). The substrate (black) is bound by the binding site to orient it next to the triad. ( PDB: 1LVM )
What are catalytic triads?
A catalytic triad is a set of three coordinated amino acids that can be found in the active site of some enzymes. Catalytic triads are most commonly found in hydrolase and transferase enzymes (e.g. proteases, amidases, esterases, acylases, lipases and β-lactamases ). An Acid - Base - Nucleophile triad is a common motif for generating a nucleophilic residue for covalent catalysis. The residues form a charge-relay network to polarise and activate the nucleophile, which attacks the substrate, forming a covalent intermediate which is then hydrolysed to release the product and regenerate free enzyme. The nucleophile is most commonly a serine or cysteine amino acid, but occasionally threonine or even selenocysteine. The 3D structure of the enzyme brings together the triad residues in a precise orientation, even though they may be far apart in the sequence ( primary structure ).
Why do amino acids deprotonate?
Since no natural amino acids are strongly nucleophilic, the base in a catalytic triad polarises and deprotonates the nucleophile to increase its reactivity. Additionally, it protonates the first product to aid leaving group departure.
Which residue aligns and polarises the base (usually histidine) which activates the nucleophil?
The acid residue (commonly glutamate or aspartate) aligns and polarises the base (usually histidine) which activates the nucleophile (often serine or cysteine, occasionally threonine ). The triad reduces the p Ka of the nucleophilic residue which then attacks the substrate.
When was chymotrypsin solved?
The structure of chymotrypsin was solved by X-ray crystallography in the 1960s , showing the orientation of the catalytic triad in the active site. Other proteases were sequenced and aligned to reveal a family of related proteases, now called the S1 family.
How do catalytic triads work?
Catalytic triads perform covalent catalysis via an acyl-enzyme intermediate. If this intermediate is resolved by water, the result is hydrolysis of the substrate. However, if the intermediate is resolved by attack by a second substrate, then the enzyme acts as a transferase. For example, attack by an acyl group results in an acyltransferase reaction. Several families of transferase enzymes have evolved from hydrolases by adaptation to exclude water and favour attack of a second substrate. In different members of the α/β-hydrolase superfamily, the Ser-His-Asp triad is tuned by surrounding residues to perform at least 17 different reactions. Some of these reactions are also achieved with mechanisms that have altered formation, or resolution of the acyl-enzyme intermediate, or that don't proceed via an acyl-enzyme intermediate.
When was trypsin first purified?
The enzymes trypsin and chymotrypsin were first purified in the 1930s. A serine in each of trypsin and chymotrypsin was identified as the catalytic nucleophile (by diisopropyl fluorophosphate modification) in the 1950s. The structure of chymotrypsin was solved by X-ray crystallography in the 1960s, showing the orientation ...
What is the name of the peptide that inhibits chymotrypsin?
5. N-Tosylamido-L-phenylethyl, also called tosyl phenylalanyl chloromethyl ketone, (TPCK) irreversibly inhibits chymotrypsin, and is used as to label the active site histidine residue. The COCH2Cl group is the reactive group that binds to the His residue.
How many steps are there in chymotrypsin catalysis?
Note that your textbook may not break out each of these steps individually, but all steps should be ordered. (There will be eight steps.)
What enzymes are involved in cleaving peptide bonds?
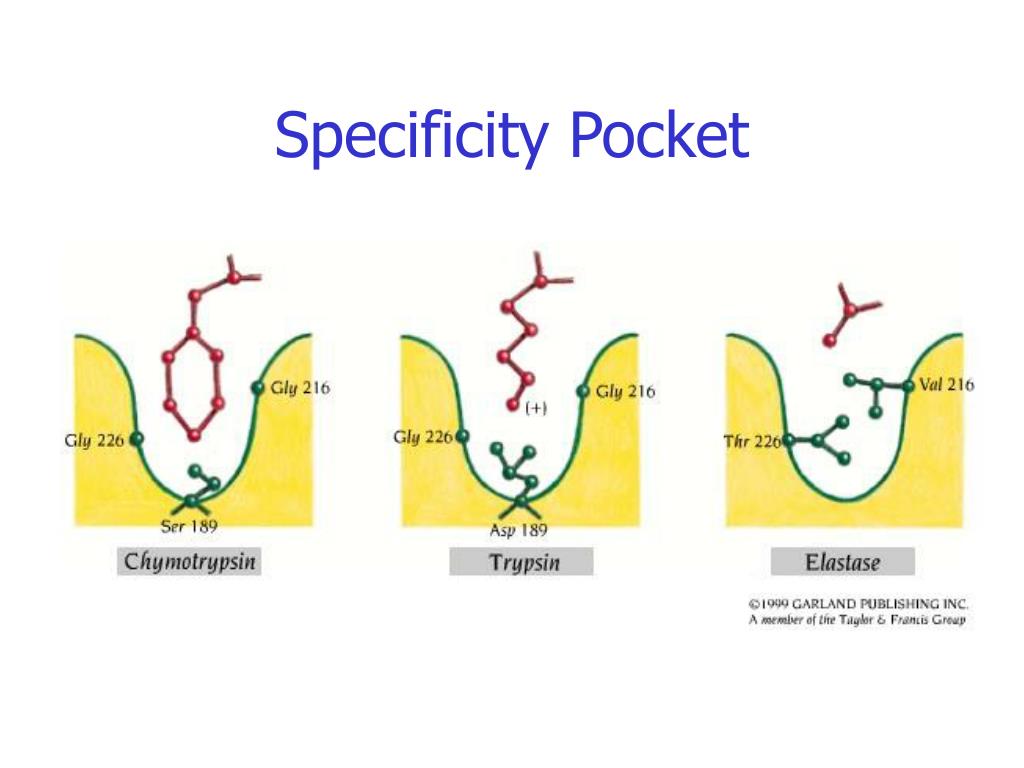
Chymotrypsin , trypsin, and elastase are digestive enzymes called serine proteases. The serine proteases differ in substrate specificity: Chymotrypsin cleaves peptide bonds after aromatic or bulky hydrophobic side chains; trypsin requires basic amino acid residues; and elastase cleaves bonds following small uncharged side chains. A chart of amino acids can be found here. The specificity pockets (substrate-binding sites) of each of the serine proteases are drawn below.
What is the phenyl group of TPCK?
The phenyl group of TPCK is structurally similar to regular chymotrypsin substrates. 5. N-Tosylamido-L-phenylethyl, also called tosyl phenylalanyl chloromethyl ketone, (TPCK) irreversibly inhibits chymotrypsin, and is used as to label the active site histidine residue.
What enzyme cleaves the P-O5 bond?
RNAse is an enzyme that cleaves the P-O5' bond in RNA. It has two His residues in the active site. Suggest a plausible explanation why the enzyme activity changes when pH is increased or decreased from ~pH 6.0, as shown in the graph below. -- explain answers.
Which amino acid is positively charged at pH 7?
Lysine and arginine both have basic amino acid side chains that are positively charged at pH 7. Trypsin's substrate-binding site contains a negatively charged amino acid residue. Elastase cleaves peptide bonds after amino acids with small side chains, such as glycine, alanine, or valine.
What peptides fit in the S1 pocket?
Leucine (Leu), isoleucine (Ile), methionine (Met), tryptophan (Trp), or phenylalanine ( Phe) could fit in the S1' pocket. The peptide residues on the amino side of the bond to be broken (the scissile bond) are labeled P1, P2, P3, etc. Residues on the carboxyl side of the scissile bond are labeled P1', P2', P3', etc.
What is the crystal structure of trypsin?
Crystal structure of Trypsin, a typical serine protease. Serine proteases (or serine endopeptidases) are enzymes that cleave peptide bonds in proteins, in which serine serves as the nucleophilic amino acid at the (enzyme's) active site. They are found ubiquitously in both eukaryotes and prokaryotes.
Where is trypsin produced?
Trypsin, a powerful digestive enzyme, is generated in the pancreas. Inhibitors prevent self-digestion of the pancreas itself. Serine proteases are paired with serine protease inhibitors, which turn off their activity when they are no longer needed.
What is the reaction of serine OH and nitrogen?
The serine -OH attacks the carbonyl carbon, and the nitrogen of the histidine accepts the hydrogen from the -OH of the [serine] and a pair of electrons from the double bond of the carbonyl oxygen moves to the oxygen. As a result, a tetrahedral intermediate is generated.
What are the inhibitors of serine proteases?
Serine proteases are inhibited by a diverse group of inhibitors, including synthetic chemical inhibitors for research or therapeutic purposes, and also natural proteinaceous inhibitors. One family of natural inhibitors called "serpins" (abbreviated from serine protease inhibitors) can form a covalent bond with the serine protease, inhibiting its function. The best-studied serpins are antithrombin and alpha 1-antitrypsin, studied for their role in coagulation / thrombosis and emphysema / A1AT, respectively. Artificial irreversible small molecule inhibitors include AEBSF and PMSF .
What is the role of host organisms in serine proteases?
Host organisms must ensure that the activity of serine proteases is adequately regulated . This is achieved by a requirement for initial protease activation, and the secretion of inhibitors.
What amino acids are involved in the oxyanion hole?
It was discovered that additional amino acids of the protease, Gly 193 and Ser 195 , are involved in creating what is called an oxyanion hole. Both Gly 193 and Ser 195 can donate backbone hydrogens for hydrogen bonding. When the tetrahedral intermediate of step 1 and step 3 are generated, the negative oxygen ion, having accepted the electrons from the carbonyl double bond, fits perfectly into the oxyanion hole. In effect, serine proteases preferentially bind the transition state and the overall structure is favored, lowering the activation energy of the reaction. This "preferential binding" is responsible for much of the catalytic efficiency of the enzyme.
What is the substrate specificity of proteases?
Serine proteases are characterised by a distinctive structure, consisting of two beta-barrel domains that converge at the catalytic active site. These enzymes can be further categorised based on their substrate specificity as either trypsin-like, chymotrypsin-like or elastase-like.

Overview
A catalytic triad is a set of three coordinated amino acids that can be found in the active site of some enzymes. Catalytic triads are most commonly found in hydrolase and transferase enzymes (e.g. proteases, amidases, esterases, acylases, lipases and β-lactamases). An acid-base-nucleophile triad is a common motif for generating a nucleophilic residue for covalent catalysis. The residues f…
History
The enzymes trypsin and chymotrypsin were first purified in the 1930s. A serine in each of trypsin and chymotrypsin was identified as the catalytic nucleophile (by diisopropyl fluorophosphate modification) in the 1950s. The structure of chymotrypsin was solved by X-ray crystallography in the 1960s, showing the orientation of the catalytic triad in the active site. Other proteases were sequenced and aligned to reveal a family of related proteases, now called the S1 family. Simulta…
Function
Enzymes that contain a catalytic triad use it for one of two reaction types: either to split a substrate (hydrolases) or to transfer one portion of a substrate over to a second substrate (transferases). Triads are an inter-dependent set of residues in the active site of an enzyme and act in concert with other residues (e.g. binding site and oxyanion hole) to achieve nucleophilic catalysis. These triad residues a…
Identity of triad members
The side-chain of the nucleophilic residue performs covalent catalysis on the substrate. The lone pair of electrons present on the oxygen or sulfur attacks the electropositive carbonyl carbon. The 20 naturally occurring biological amino acids do not contain any sufficiently nucleophilic functional groups for many difficult catalytic reactions. Embedding the nucleophile in a triad increases its r…
Examples of triads
The Serine-Histidine-Aspartate motif is one of the most thoroughly characterised catalytic motifs in biochemistry. The triad is exemplified by chymotrypsin, a model serine protease from the PA superfamily which uses its triad to hydrolyse protein backbones. The aspartate is hydrogen bonded to the histidine, increasing the pKa of its imidazole nitrogen from 7 to around 12. Thi…
Divergent evolution
The sophistication of the active site network causes residues involved in catalysis (and residues in contact with these) to be highly evolutionarily conserved. However, there are examples of divergent evolution in catalytic triads, both in the reaction catalysed, and the residues used in catalysis. The triad remains the core of the active site, but it is evolutionarily adapted to serve …
Convergent evolution
The enzymology of proteases provides some of the clearest known examples of convergent evolution. The same geometric arrangement of triad residues occurs in over 20 separate enzyme superfamilies. Each of these superfamilies is the result of convergent evolution for the same triad arrangement within a different structural fold. This is because there are limited productive ways to ar…
See also
• Active site
• Convergent evolution
• Divergent evolution
• Enzyme catalysis
• Enzyme superfamily