What is the function of neuraminidase spikes (N spikes)? help the virus separate from infected cells after replication via enzyme neuraminidase Flu transmission aerosol Flu symptoms chills, fever, headache, general muscular aches (NO gastrointestinal symptoms)
What is the difference between hemagglutinin spikes and neuraminidase spikes?
Each hemagglutinin spike is composed of a trimer of three molecules, while the neuraminidase spike consists of a tetramer (Figure 4.9 ). The hemagglutinin spikes are responsible for binding the influenza virus receptor, which is sialic acid ( N -acetyl neuraminic acid), a sugar group commonly found on a variety of glycosylated molecules.
What is the function of neuraminidase?
Neuraminidase is a membrane glycoprotein enzyme that cleaves sialic acid, helping progeny influenzaviruses leave without reinfecting the host cell. Like hemagglutinin, each neuraminidase molecule consists of a globular head on a stalk, extending out to about 120 Å from the membrane.
How does neuraminidase activity affect progeny virus?
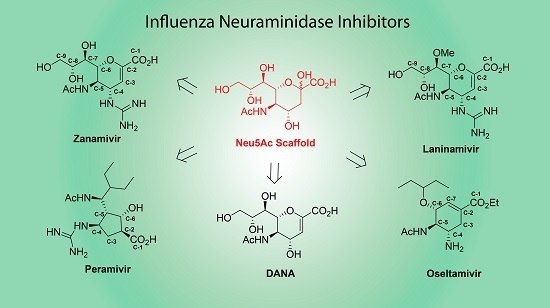
A small molecule that inhibits the neuraminidase activity will block the release of newly assembled particles and thereby progeny virus will not spread to neighboring cells. Two drugs with such activity were developed upon the approval of the FDA. Oseltamivir and zanamivir are analogs of sialic acid residue, which is a substrate for neuraminidase.
What is the role of neuraminidase in the pathogenesis of influenza type a?
Influenza type A virus neuraminidase does not play a role in viral entry, replication, assembly, or budding. J. Virol. 69, 1099–1106.
See more

What are neuraminidase spikes?
Neuraminidases are enzymes that cleave sialic acid (also called neuraminic acid) groups from glycoproteins. Neuraminidase inhibitors are antiviral agents that inhibit influenza viral neuraminidase activity and are of major importance in the control of influenza.
What is the role of neuraminidase?
Influenza A viruses generally mediate binding to cell surface sialic acid receptors via the hemagglutinin (HA) glycoprotein, with the neuraminidase (NA) glycoprotein being responsible for cleaving the receptor to allow virus release.
What is the function of N spikes on an influenza virus?
The N spikes are an enzyme called neuraminidase. N spikes help break down the mucous material surrounding host cells in the respiratory tract and initiate penetration of the virus into the host cell. These spikes are not the same in all influenza A viruses.
What is the function of neuraminidase in virus?
It is believed that the major function of viral neuraminidase (NA) is at the final stage of infection when NA cleaves sialic acid from cell surface and progeny virions facilitating virus release from infected cells (1, 2).
What do NA spikes of influenza bind to?
The hemagglutinin spikes are responsible for binding the influenza virus receptor, which is sialic acid (N-acetyl neuraminic acid), a sugar group commonly found on a variety of glycosylated molecules.
What is the function of the hemagglutinin spikes of the flu virus?
The hemagglutinin(HA) of influenza virus is a major glycoprotein and plays a crucial role in the early stage of virus infection: HA is responsible for binding of the virus to cell surface receptors, and it mediates liberation of the viral genome into the cytoplasm through membrane fusion.
What role do H Spikes play in influenza A infections quizlet?
What role do H spikes play in influenza A infections? They identify a specific glycoprotein embedded in the membrane of a host cell.
What is a spike protein in a virus?
The spike protein is found on the surface of the virus that causes COVID-19. After the protein piece is made, our cells break down the mRNA and remove it, leaving the body as waste. Next, our cells display the spike protein piece on their surface. Our immune system recognizes that the protein does not belong there.
What is the role of N neuraminidase proteins of influenza viruses?
By far, the most characterized function of NA is its action as a sialidase enzyme, enabling release of new virion progeny by enzymatically cleaving sialic acids from cell surface receptors and from carbohydrate side chains on nascent virions (Gottschalk, 1958; Palese et al., 1974).
What is the function of hemagglutinin and neuraminidase?
Hemagglutinin-neuraminidase (HN) protein, which is responsible for virus attachment, interacts with the fusion protein in a virus type-specific manner to induce efficient membrane fusion.
What do neuraminidase inhibitors do?
Neuraminidase inhibitors block the function of the viral neuraminidase protein, thus stopping the release of viruses from the infected host cells and preventing new host cells from being infected, and therefore, the infection does not spread in the respiratory tract.
What type of enzyme is neuraminidase?
glycoside hydrolase enzymesNeuraminidase, also known as sialidase, is a group of glycoside hydrolase enzymes that cleave the glycosidic linkages of neuraminic acids.
What is the function of hemagglutinin and neuraminidase?
Hemagglutinin-neuraminidase (HN) protein, which is responsible for virus attachment, interacts with the fusion protein in a virus type-specific manner to induce efficient membrane fusion.
What do neuraminidase inhibitors do?
Neuraminidase inhibitors block the function of the viral neuraminidase protein, thus stopping the release of viruses from the infected host cells and preventing new host cells from being infected, and therefore, the infection does not spread in the respiratory tract.
What type of enzyme is neuraminidase?
glycoside hydrolase enzymesNeuraminidase, also known as sialidase, is a group of glycoside hydrolase enzymes that cleave the glycosidic linkages of neuraminic acids.
What role do hemagglutinin and neuraminidase play when a flu virus attacks a human cell?
The influenza virus major surface glycoproteins, hemagglutinin (HA), and neuraminidase (NA) dominate the virion surface and form the main targets for these neutralizing antibodies. In addition to the mutations that arise due to antigenic drift, the HA and NA of influenza A viruses (IAVs) can exist in different forms.
What is the role of neuraminidase in the release of viral particles?
The neuraminidase activity of NA protein plays a major role in the release of nascent viral particles assembled in infected cells (see Fig. 15.2 ). It facilitates the release of viral particles from cells by the cleavage of a sialic acid residue of glycan moiety linked to cellular glycoproteins in the plasma membrane. A small molecule that inhibits the neuraminidase activity will block the release of newly assembled particles and thereby progeny virus will not spread to neighboring cells. Two drugs with such activity were developed upon the approval of the FDA. Oseltamivir and zanamivir are analogs of sialic acid residue, which is a substrate for neuraminidase. In particular, osetamivir was aptly used during the 2009 H1N1 pandemic.
What are neuraminidase inhibitors?
In some cases, neuraminidases act as a virulence factors.69 Thus, the inhibitor of virus neuraminidase is regarded as a potential agent with antiviral and antibacterial activities. It has been reported that several polymer-based inhibitors of influenza virus are multivalent in neuraminic acid A (NeuAc). 8,35,70,71 The synthetic random co-polymers bearing NeuAc via C-glycoside linkage have been shown to inhibit the agglutination of erythrocyte by influenza viruses in vitro. 72 For the synthesis of C-glycoside polymer, an enzymatic approach was reported by Linhardt. 73 An NeuAc monomer with phenolic aglycon was polymerized with soybean peroxidase to afford a molecular weight of 20 000, which is similar to MG2 mucin (MUC7). MG2 contains relatively homogeneous sialylated disaccharides and trisaccharides and binds to a wide variety of microorganisms. The resulting C-glycoside polymer was investigated for the inhibitory effect against neuraminidase from Clostridium perfringens by using 4MU-labeled fluorescent substrate to give a Ki value of 900 nM based on the polymer concentration. The observed Ki was over 10-fold lower than the monomeric C-glycoside of NeuAc.
What enzyme removes terminal sialic acids from gangliosides?
Neuraminidase (sialidase) removes terminal sialic acids from gangliosides. Using this technique it has been concluded that all GM3 was present on the membrane surface of the enveloped Sindbis virus.58,59 In neuroblastoma cells, neuraminidase from Vibrio cholerae cleaved the sialic acid from most GM3, while it also removed the terminal sialic acid from GD1a to yield GM1a. 60 In contrast, in macrophages most GD1a but no GM3 was degraded by this enzyme. 61 Neuraminidase from Clostridium perfringens was used to study transport of newly synthesized GM3, GD3, and GT3 to the surface of retina cells. 62
What is the neuraminidase produced by?
Neuraminidase is produced by various mucosal pathogens and is considered a virulence factor in that it modifies the host's response to infection. C. diphtheriae produce neuraminidase, which cleaves N -acetylneuraminic acid (NAN) from cell surfaces to produce pyruvate (a growth stimulant). Neuraminidases have different requirements for divalent cations. There are conflicting reports for C. diphtheriae neuraminidase's cation requirement, so this remains an open question. Neuraminidase is produced from many strains of C. diphtheriae, regardless of DT expression. Neuraminidase enzymatic activity is also independent of sucrose fermentation, isolation site, and hemagglutination. Strains with neuraminidase activity are able to agglutinate human erythrocytes with high titers. Neuraminidase is found inside the cell, on the surface, and as a free soluble mediator. After extensive subculture in vitro, some species lose or have drastically reduced production of neuraminidase, which may suggest a role for neuraminidase in vivo but not in vitro. The gene (s) encoding neuraminidase has not been identified.
Why was C. diphtheriae neuraminidase discovered?
The C. diphtheriae neuraminidase was discovered because of its presence in DT purifications. In fact, commercial diphtherial antitoxin includes diphtherial neuraminidase antibodies. Although DT and neuraminidase have similar molecular weights and stay associated during column chromatography, they are immunochemically and electrophoretically distinct. Neuraminidase production is enhanced in high-iron conditions that are inhibitory to DT production. Further, neuraminidase synthesis is unrelated to the lysogenic character of the strains. Relevant virulence factors of C. diphtheriae have been reviewed ( Ott & Burkovski, 2014 ).
What is NA inhibitor?
NA inhibitors are chemical analogs of a sialic acid. Oseltamivir is also called by trade name, Tamiflu.
Do neuraminidase have carbohydrate binding domains?
Among bacterial neuraminidases, some are known to have one or more carbohydrate-binding domains in addition to a catalytic domain. 74,75 The neuraminidases with additional lectin domains were revealed to hydrolyze multivalent substrates with much greater efficiency than the monovalent substrate by Boons et al. 76 The multivalent substrate exhibited 100-fold smaller Km than the monovalent derivative. It seems that the catalytic and lectin domains interact simultaneously with the multivalent substrates, leading to the affinity enhancement. Furthermore, the catalytic activity of neuraminidases was effectively inhibited by the multivalent polymer displaying galactose, a ligand for the lectin domain. The Ki of the galactoside polymer against neuraminidase from Vibrio cholerae is 50 μ M, which is 100-fold smaller than that of monovalent d -galactose. The polymer did not inhibit the neuraminidase from Salmonella typhimurium, which does not have a lectin domain. This new concept of inhibitor that targets lection domains of the enzyme has a great potential to develop a specific inhibitor for bacterial neuraminidase.
What is the disease associated with Reyes syndrome?
d. Infection with influenza and treatment with aspirin are associated with the development of Reyes syndrome.
How to treat rabies bites?
a. Immediately wash the wound with soap and water, inject the bite area with rabies immunoglobulin, and begin a series of four rabies vaccine injections.
Which type of drift has produced major antigenic changes in the viral genome?
a. Antigenic drift has produced major antigenic changes in the viral genome.
What is the neuraminidase?
Neuraminidase is an exosialidase (EC 3.2.1.18) which cleaves α-ketosidic linkage between the sialic (N-acetylneuraminic) acid and an adjacent sugar residue [1]. The amino acid sequence of NA is coded by the 6th RNA segment. Nine subtypes of NA are described for influenza A, whereas only one NA subtype was revealed for the influenza viruses B and C [2]. Nine subtypes of influenza A NA are divided into two phylogenic groups. The first group consists of the neuraminidases of N1, N4, N5 and N8 subtypes, and the second one consists of N2, N3, N6 N7 and N9 subtypes [3].
What is the second neuraminic acid binding site?
The second neuraminic acid binding site, the so called HBsite, was found in N9 neuraminidases [10]. The a.a. sequence of this site is highly conservative among avian influenza viruses. This site is formed by three NA loops:
What is the function of the HB site?
The function of the HB-site has yet to be clarified. It has been suggested that it may play the role of an alternative neuraminic acid binding site, in other words , function as a surrogate of the influenza virus HA; this assumption is based on the existence of viruses with combined HA and NA functions in one protein molecule, such as the ND virus. The HB-site, described earlier, is common for the NAs of viruses which HA interacts with α2-3-sialylated carbohydrate chains (i.e. avian and equine influenza viruses), at the same time, the key amino acid positions of this site are changed in viruses with α2-6-specificity (human, swine, and poultry H9N2 viruses). It is worth mentioning that the H9N2 influenza viruses isolated from poultry in Hong Kong and viruses of H2N2 and H3N2 subtypes, which caused human pandemics, have similar changes in the HB-site sequence. This data allows one to suggest that some species of poultry may act as intermediate hosts in the influenza virus transfer from its natural reservoir (waterfowl population) to humans [11].
What is the structure of the cytoplasmic, transmembrane and stem domains?
The three-dimensional structure of cytoplasmic, transmembrane and stem domains has not been determined yet (due to the features of the enzymes, which are used for cleavage of this membrane protein from the virion, the crystallized region starts at residues ~74-77) [6]. There is speculation about the presence of an α-helix motif in the uncrystallized structure, which has been supported by cryoelectron microscopy [4]. Therefore, one can unequivocally judge only the chain folding of the head region of the enzyme (as part of the tetrameric structure). The NA's head region consists of one big domain, which is formed by six identical antiparallel β-sheets (motifs) organized in the form of a propeller-like structure. Loops connecting the motifs and loops between every second and third strain of each motif are of ultimate importance for the enzyme [2]. Loops are the most variable parts of the structure of all NAs; they vary in length and can even have some arranged elements typical of the secondary structure. For example, loops of N9 NA have some α-helix regions: residues 106 – 110 form one spiral turn (α), located above the С-terminus of the polypeptide chain, which consecutively forms another α-helix turn, and a.a. 144 – 146 of the neighboring subunit forms the (310) helix. The 310 helix and two chains (106 − 110 and C-terminus) form the antiparallel layer [6]. The loop connecting the fourth and the fifth motifs is the longest one and is stabilized by a disulfide bridge located between Cys318 and Cys337; it also has the conservative ion pair Asp330-Arg364 and the Ca2+binding site [1].
Which enzyme promotes the O-deacetylation of the N-acetyl-9-O-?
The enzyme of the influenza C virus does not belong to the neuraminidase group. It promotes the O-deacetylation of the N-acetyl-9-O-acetylneuraminic acid, i.e. it belongs to the esterase family and will not be considered in this review.
Does zanamivir cause resistance?
Until recently, it was considered that active uncontrolled use of zanamivir and oseltamivir would not have a significant influence on the development of resistance in influenza virus strains. That is, even if resistant strains emerge, they would not be able to replicate in the absence of the inhibitor [24]. The number of resistant viruses isolated in clinical trials accounted for less than 1%, same as their presence among seasonal influenza virus isolates worldwide.
Where is the calcium binding site located?
The calcium binding site, which is located inside the molecule (particularly under the active site, if it is placed in accordance with the picture provided) is formed by the oxygen of the main chain residues 297, 345 and 348, as well as by the oxygen of the side chain of Asp324 [1, 6]. Additionally, this site is formed by a.a. 293, 347, 111-115 and 139-143 [8].