What begins the process of transcription?
Transcription begins when RNA polymerase binds to a promoter sequence near the beginning of a gene (directly or through helper proteins). RNA polymerase uses one of the DNA strands (the template strand) as a template to make a new, complementary RNA molecule. Transcription ends in a process called termination.
What are the stages of transcription?
The process of transcription is enzymatically catalysed into three steps:
- Initiation
- Elongation
- Termination
What is the termination sequence?
terminator sequence. A sequence in DNA that signals termination of transcription to RNA Polymerase. This should not be confused with terminator codons that are the stopping signal for translation. Also known as: terminator, rho -independent termination site.
How do you transcribe DNA sequence?
- The DNA helix unravels, and the unravelling is catalysed by the enzyme helicase.
- From the two unravelled strands, one of them is used as a template.
- The exposed bases on the template attract free nucleotides from the nucleus
- The free nucleotides are joined together in a polymerisation reaction catalysed by the enzyme polymerase.
Why is transcription important?
Transcription is an essential step in using the information from genes in our DNA to make proteins. Proteins are the key molecules that give cells structure and keep them running. Blocking transcription with mushroom toxin causes liver failure and death, because no new RNAs—and thus, no new proteins—can be made.
What is the process of copying a gene's DNA sequence to make an RNA molecule?
Transcription is the process in which a gene's DNA sequence is copied (transcribed) to make an RNA molecule.
What is the process of RNA polymerase?
Transcription ends in a process called termination. Termination depends on sequences in the RNA, which signal that the transcript is finished.
How does RNA polymerase open up DNA?
Once the RNA polymerase has bound, it can open up the DNA and get to work . DNA opening occurs at the element, where the strands are easy to separate due to the many As and Ts (which bind to each other using just two hydrogen bonds, rather than the three hydrogen bonds of Gs and Cs).
Which end of the RNA strand does RNA polymerase add a matching nucleotide?
For each nucleotide in the template, RNA polymerase adds a matching (complementary) RNA nucleotide to the 3' end of the RNA strand. [See the chemical reaction] RNA polymerase synthesizes an RNA transcript complementary to the DNA template strand in the 5' to 3' direction.
Which direction does RNA polymerase read DNA?
It synthesizes the RNA strand in the 5' to 3' direction, while reading the template DNA strand in the 3' to 5' direction. The template DNA strand and RNA strand are antiparallel.
What is the first step in gene expression?
Transcription is the first step of gene expression. During this process, the DNA sequence of a gene is copied into RNA. Before transcription can take place, the DNA double helix must unwind near the gene that is getting transcribed. The region of opened-up DNA is called a transcription bubble.
Which sequences signal the cell when to turn on the gene?
Figure 9a. The promoter has DNA sequences that signal the cell when to turn on the gene and where to start reading the gene information. (image by d. Namuth-Covert)
What is the mRNA made from transcription of the ALS gene?
Each time an RNA polymerase goes through the process, one copy of the RNA is made by reading the DNA template .The type of RNA made from transcription of the ALS gene is called a messenger RNA or mRNA for short. Some
What is the function of the N promoter?
The#N#promoter#N#A specific DNA sequence to which RNA polymerase binds and initiates transcription. This region contains information which regulates when and how often the gene is transcribed and ultimately the amount of protein it produces.#N#’s role is to selectively turn on the ALS gene in cells that will need the ALS enzyme. The promoter accomplishes this by its specific nucleotide sequences. We will discuss the regulation of gene expression in more detail later. What cells in a plant will need the ALS#N#enzyme#N#A protein that catalyzes, or speeds up, a specific biochemical reaction without changing the nature of the reaction.#N#? All cells that are making proteins. Obviously this will be all cells in the plant at some stage of development. Therefore the ALS#N#gene#N#The fundamental unit of heredity that carries genetic information from one generation to the next. A gene is an ordered sequence of nucleotides located on a particular position on a particular chromosome that encodes a specific functional protein.#N#has a promoter sequence that allows the gene to be turned on in all types of cells.
What is the chromosome?
chromosome. A genetic structure in a cell composed of condensed DNA, which contains the genetic code for an organism. might be expressed in the wrong cells or wrong times. Figure 10. RNA polymerase encounters the TATA box at the end of the promoter. This signals where to begin the reading of the coding region .
Where is the tata box in RNA polymerase?
Figure 10. RNA polymerase encounters the TATA box at the end of the promoter. This signals where to begin the reading of the coding region . (Image by D. Namuth-Covert)
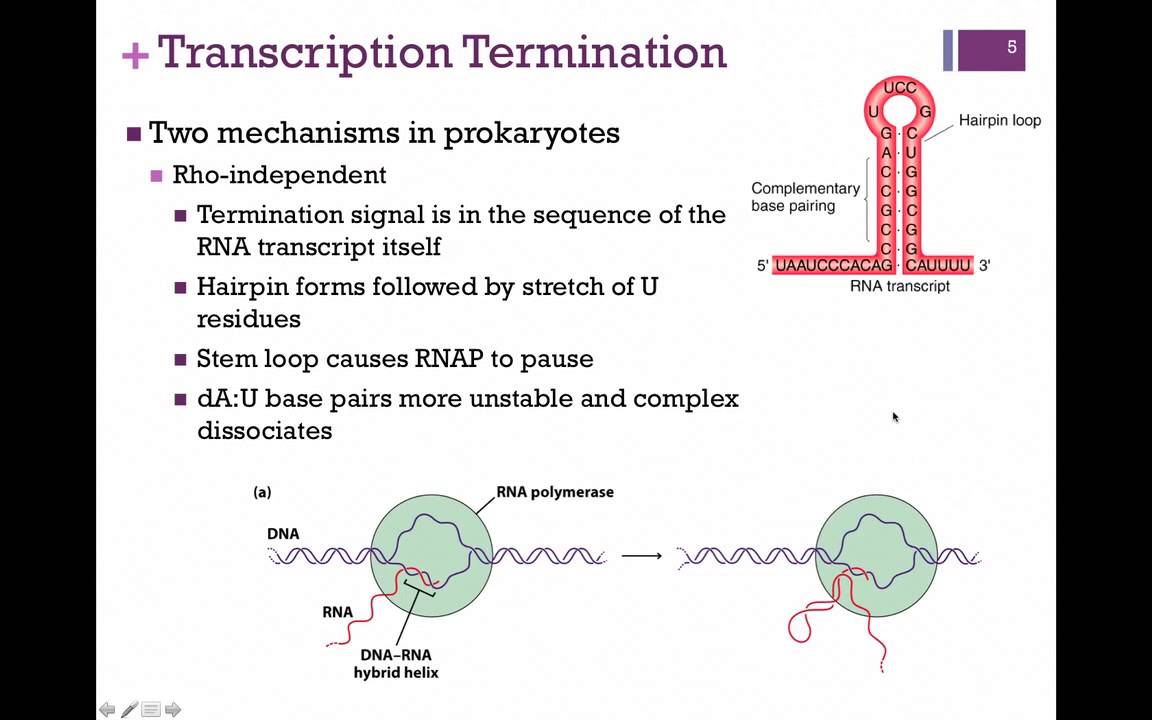
Is the termination sequence palindromic?
Figure 13. The termination sequence is palindromic. The RNA being made has a sequence that can form a hairpin loop. (Image by D. Lee)
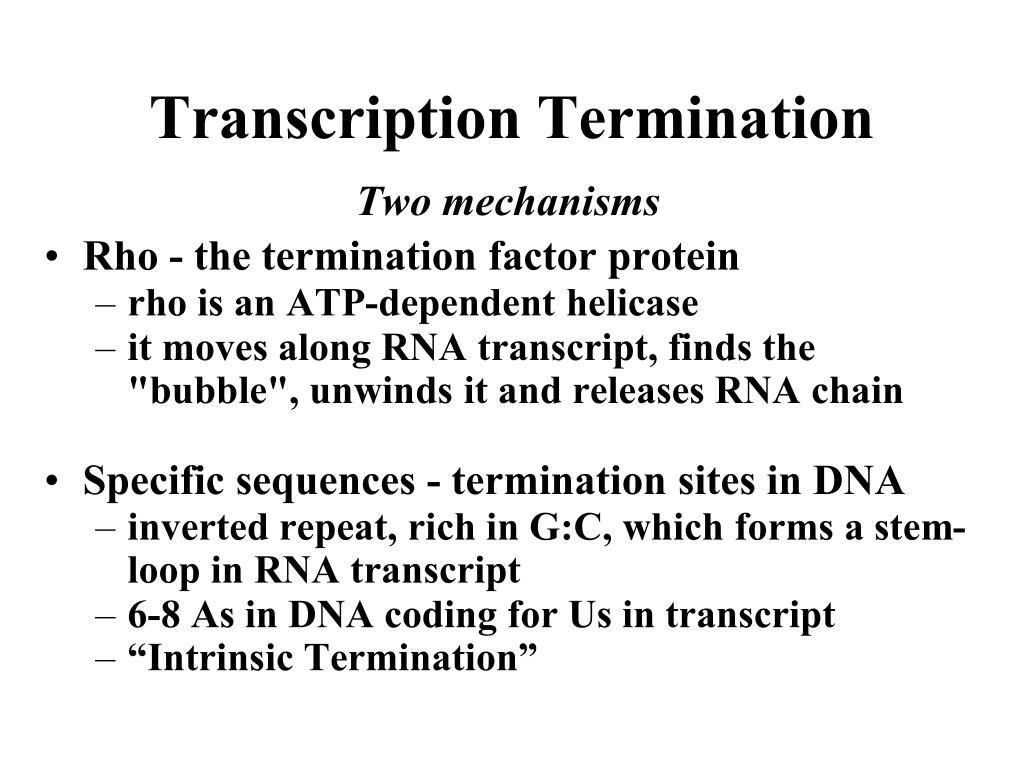
What is the termination of transcription?
Specific base sequences signal the termination of transcription. The signal may be described as an inverted-repeat sequence which contains an intervening non-repeating segment (Figure 17.4 ). Transcription cannot terminate at an exclusive sequence since the З’-end of transcripts initiated from a single promoter may vary in the numbers of consecutive Us and may include other bases. The last transcribed base is always 27–43 bases from the start of the termination sequence. The actual mechanism of termination has not been fully elucidated but it is believed that termination occurs through sequence-promoted pausing of RNA polymerase activity. The first pause may be caused by the inverted-repeat sequence and the second pause by the high G + C region. The pauses may be the consequence of transcript folding. Termination in some cases involves additionally the presence of a termination protein called ρ (rho). On binding tightly to segments containing repeating Cs, ρ acquires an ATPase activity upon which its role depends. The final event in the termination of transcription is the dissociation of the core enzyme from the DNA. On its interaction with any free σ subunit, the holoenzyme is reconstituted and may initiate the transcription of another gene.
How does Rho factor precipitate termination of transcription?
Rho factor precipitates termination of transcription by first binding to a site on the nascent transcript, then using its ATP hydrolysis activity as a source of energy to dissociate the transcript from its ternary complex with RNA polymerase and DNA.
How does an intrinsic terminator work?
An intrinsic terminator can be sensitive to RNA rearrangements when sequences that participate in the formation of the terminator helix can be sequestered into an alternate RNA structure designated an antiterminator. The decision between formation of the mutually exclusive terminator and antiterminator structures can be determined by binding of an extrinsic factor; in riboswitch systems, the RNA structure is modulated by direct binding of an effector molecule (such as a small molecule or RNA). Binding of effector molecules can promote the formation of the terminator (resulting in inhibition of transcription of the downstream coding sequences) or the antiterminator (resulting in increased gene expression). In systems in which the terminator form of the RNA is more stable than the antiterminator form, the expression is off in the absence of effector binding, so that the effector acts as an inducer to increase the expression of the downstream gene (s) ( Figure 1 (b) ). If the antiterminator is more stable than the terminator, then expression is on in the absence of the effector, which serves as a repressor molecule ( Figure 1 (c) ). In this case, effector binding often stabilizes an element that includes sequences that would otherwise form the 5′ side of the antiterminator structure; this alternate element serves as an anti-antiterminator, as it competes with antiterminator formation, and stabilization of the anti-antiterminator prevents the formation of the more stable antiterminator and allows the less stable terminator helix to form ( Figure 1 (d) ).
What is the transcriptional unit of genes?
The transcriptional unit of genes regulated at this level includes a transcriptional terminator (or attenuator), positioned upstream of the regulated coding sequence (s). This arrangement results in two classes of transcripts originating from a single promoter, a shorter transcript resulting from termination at the attenuator site and a longer transcript resulting from transcription elongation past the attenuator ( Figure 1 (a) ). The regulated attenuator is usually an intrinsic terminator capable of functioning in the absence of dedicated termination factors such as Rho protein. Intrinsic terminators are active within the nascent RNA transcript, and they consist of a G + C-rich helix immediately followed by a U-run that forms a U-A RNA–DNA helix within RNA polymerase (RNAP). Folding of the nascent RNA into the helix, while RNAP is paused at the end of the U-run, affects the stability of the transcription elongation complex and results in termination of transcription and release of RNAP and the terminated transcript from the DNA template.
What is required for 3′ end processing?
For efficient 3′ end processing, two separate sequence elements are required: the AATAAA sequence and a second GT-rich element immediately downstream of the cleavage site at which poly (A) is added. Although 3′ processing may initiate the termination process, actual termination is believed to occur at the RNA Pol II pause site (s). The pause site may be a hairpin structure and/or a sequence element that binds a specific protein factor. One such termination signal has been identified to be the CCAAT box sequence which functions as an orientation-dependent, position-independent terminator ( 99 ). Most probably the termination signal is mediated by the specific-binding protein CP1, similar to the role supposedly played by an AATAAA-binding factor (s).
What is the function of sigma in RNA polymerase?
RNA polymerase needs the sigma subunit to recognize and bind to the promoter. Once RNA polymerase moves forward and starts transcribing, it releases sigma and picks up NusA protein. After termination, NusA protein is displaced by sigma.
What happens when RNAP is paused?
Folding of the nascent RNA into the helix, while RNAP is paused at the end of the U-run, affects the stability of the transcription elongation complex and results in termination of transcription and release of RNAP and the terminated transcript from the DNA template. Sign in to download full-size image.