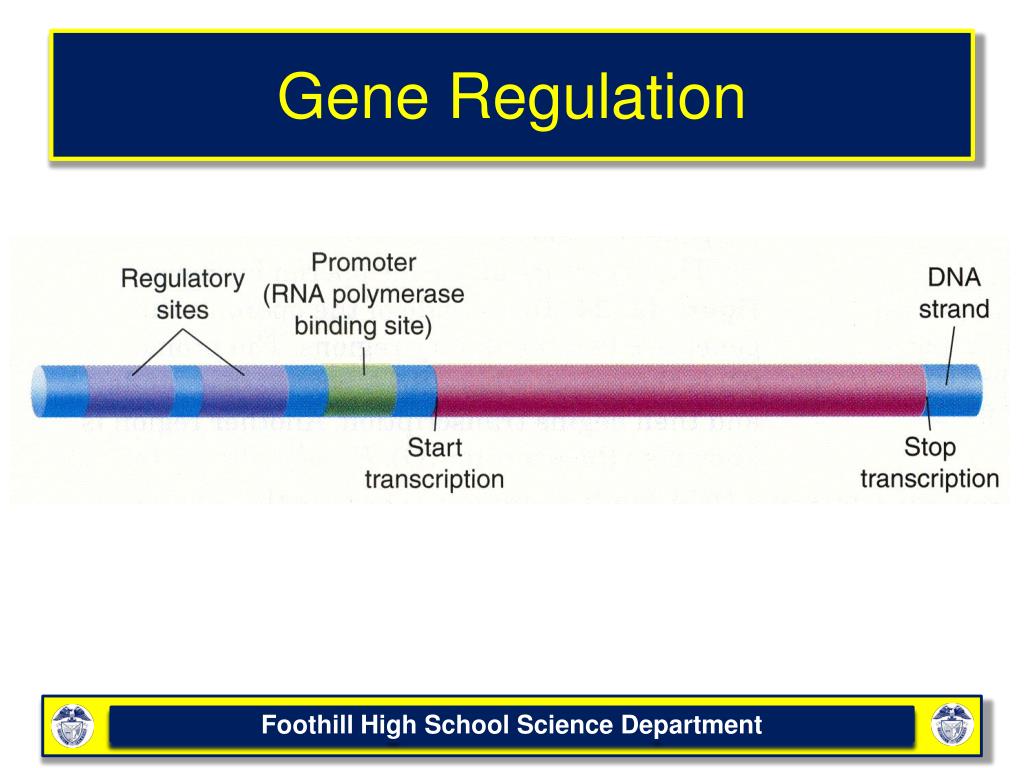
There are three broad levels of regulating gene expression:
- transcriptional control (whether and how much a gene is transcribed into mRNA)
- translational control (whether and how much an mRNA is translated into protein)
- post-translational control (whether the protein is in an active or inactive form, and whether the protein is stable or degraded)
Full Answer
What is a regulatory element in biology?
A regulatory element is a DNA sequence that certain transcription factors recognize and bind to in order to recruit or repel RNA polymerase. The promoter along with nearby transcription factor binding elements regulate gene transcription. Regulatory elements can be used for either positive and negative transcriptional control.
What are the primary regulatory components of the genome?
Regulatory elements including promoters and enhancers are the primary regulatory components of the genome. The regulatory region can be divided into cis-regulatory elements and trans-regulatory elements.
What are the different types of enhancing gene regulatory elements?
We find here that in addition to these elements, two other types of enhancing gene regulatory elements exist: chromatin-dependent enhancers and closed-chromatin enhancers (Fig. 7d ). Chromatin-dependent enhancers are characterized by forkhead motifs, binding of Mediator and p300 protein and a strong signal for H3K27 acetylation.
How are genes regulated in multicellular organisms?
Gene expression changes, the driving forces for cellular diversity in multicellular organisms, are regulated by a diverse set of gene regulatory elements that direct transcription in specific cells. Mutations in these elements, ranging from chromosomal aberrations to single-nucleotide polymorphisms, are a major cause of human disease.
What are regulatory elements in genes?
A regulatory element is a DNA sequence that certain transcription factors recognize and bind to in order to recruit or repel RNA polymerase. The promoter along with nearby transcription factor binding elements regulate gene transcription.
What are the types of regulatory elements?
There are several types of gene regulatory elements, including promoters, enhancers, silencers, and insulators (Figure 1). Promoters are located near a gene's transcription start site and are where the transcription machinery assembles (for an in-depth review of promoters, see 104, 110).
What are the 3 elements of gene expression?
Regulation of transcription can be broken down into three main routes of influence; genetic (direct interaction of a control factor with the gene), modulation interaction of a control factor with the transcription machinery and epigenetic (non-sequence changes in DNA structure that influence transcription).
What are examples of regulatory genes?
An example of a regulator gene is a gene that codes for a repressor protein that inhibits the activity of an operator (a gene which binds repressor proteins thus inhibiting the translation of RNA to protein via RNA polymerase).
Where are the regulatory genes located?
They are present upstream near the transcription start sites of genes in between the operator and structural gene. Regulatory gene regulates the expression of structural genes by its protein products that are mostly transcription factors.
Where is the regulatory region of a gene?
The 5′ untranslated region (UTR) is a regulatory region of DNA situated at the 5′ end of all protein-coding genes that is transcribed into mRNA but not translated into protein. 5′UTRs contain various regulatory elements (Fig. 1b) and play a major role in the control of translation initiation.
How are genes regulated?
Gene regulation Transcription factors bind to regulatory regions of the gene and increase or decrease the level of transcription. Other mechanisms of gene regulation include regulating the processing of RNA, the stability of mRNA and the rate of translation.
What is the difference between gene expression and gene regulation?
Gene expression refers to the process by which the instructions in our DNA are converted into a functional product, such as a protein, while gene regulation refers to the process involved in turning genes on and off to ensure the appropriate expression of genes at the proper times.
What are the two functions of gene regulatory proteins?
Activators are regulatory proteins that promote transcription by enhancing the interaction of RNA polymerase with the promoter. Repressors are regulatory proteins that prevent transcription by impeding the progress of RNA polymerase along the DNA strand so the DNA cannot be transcribed to mRNA.
What do regulatory genes produce?
genetic regulation operon is controlled by a regulator gene, which produces a small protein molecule called a repressor. The repressor binds to the operator gene and prevents it from initiating the synthesis of the protein called for by the operon.
What is the difference between regulatory and structural genes?
Structural genes are those genes that code all the proteins in a genome except regulatory genes. Regulatory genes are those genes that code for proteins or factors that control the expression of structural genes. In prokaryotes, structural genes are present in a sequence called operon.
How many types of regulatory proteins are there?
two typesBroadly, two types of regulators are distinguished: enhancers increase the probability that a given gene is expressed, inhibitors decrease it. Transcription factors regulate the presence of structural proteins needed to build and maintain an organism.
What are regulatory factors?
a chemical secretion of the HYPOTHALAMUS, associated with blood sugar levels, that controls the secretion of hormones. The secretion and inhibition of hormones involve negative feedback systems (see FEEDBACK MECHANISM).
What are the two main parts of regulatory in life sciences?
Two-component regulatory systems, comprising sensor kinase and response regulator proteins, carry out signal transduction in prokaryotic and eukaryotic microorganisms, as well as plants. Response regulators act as phosphorylation-mediated switches, turning on and off cellular responses to environmental stimuli.
What are upstream regulatory elements?
Distal (upstream) regulatory elements, which can include enhancers, silencers, insulators, and locus control regions, can be located up to 1 Mb pairs from the promoter. These distal elements may contact the core promoter or proximal promoter through a mechanism that involves looping out the intervening DNA.
What are the regulatory systems?
Regulatory System means the body of legal requirements for Good Manufacturing Practices, inspections, and enforcements that ensure public health protection and legal authority to assure adherence to these requirements.
How are co-regulated genes organized?
In prokaryotes, co-regulated genes are often organized into an operon , where two or more functionally related genes are transcribed together from a single promoter into one long mRNA. This mRNA is translated to make all of the proteins encoded by the genes in the operon.
What are the levels of gene expression?
There are three broad levels of regulating gene expression: transcriptional control (whether and how much a gene is transcribed into mRNA) translational control (whether and how much an mRNA is translated into protein ) post-translational control (whether the protein is in an active or inactive form, and whether the protein is stable or degraded) ...
What are the genes that encode lactose?
The lac operon of E. coli has 3 structural genes required for metabolism of lactose, a disaccharide found at high levels in milk: 1 lacZ encodes the enzyme beta-galactosidase, which cleaves lactose into glucose and galactose 2 lacY encodes permease, a membrane protein for facilitated diffusion of lactose into the cell 3 lacA encodes transacetylase, an enzyme that modifies lactose
What are the three domains of life?
All three domains of life use positive regulation (turning on gene expression), negative regulation (turning off gene expression), and co-regulation (turning multiple genes on or off together) to control gene expression, but there are some differences in the specifics of how these jobs are carried out between prokaryotes and eukaryotes.
Why are proteins needed together?
Often a set of proteins are needed together to respond to a certain stimulus or carry out a certain function (for example, many metabolic pathways). There are often mechanisms to co-regulate such genes such that they are all transcribed in response to the same stimulus.
Do eukaryotic genes have operons?
With a few exceptions ( C. elegans and related nematodes), eukaryotic genomes do not have genes arranged in operons. Instead, eukaryotic genes that are co-regulated tend to have the same DNA regulatory element sequence associated with each gene, even if those genes are located on completely different chromosomes.
Can a gene be subject to both positive and negative transcriptional control?
A single gene can be subject to both positive and negative transcriptional control by different transcription factors, creating multiple layers of regulation. Some genes are not subject to regulation: they are constitutively expressed, meaning they are always transcribed.
Abstract and Figures
While the annotation and functional characterization of the 2% of our genome that encodes for protein has been extremely successful, the remaining 98% still remains primarily uncharted territory. Within this territory reside gene regulatory sequences that instruct genes when, where, and at what levels to turn on or off.
References (97)
Nucleotide changes in gene regulatory elements can have a major effect on interindividual differences in drug response. For example, by reviewing all published pharmacogenomic genome-wide association studies, we show here that 96.4% of the associated single nucleotide polymorphisms reside in noncoding regions.
Abstract
DNA can determine where and when genes are expressed, but the full set of sequence determinants that control gene expression is unknown. Here, we measured the transcriptional activity of DNA sequences that represent an ~100 times larger sequence space than the human genome using massively parallel reporter assays (MPRAs).
Main
The temporal and spatial pattern of gene expression is encoded in the DNA sequence; this information is read and interpreted by TFs, which recognize and bind specific short DNA sequence motifs 1. Major efforts have been undertaken to determine the DNA-binding specificities of TFs in vitro 2, 3, 4, 5 and map their binding positions in vivo 6, 7.
Results
To systematically characterize the sequence determinants of human gene regulatory element activity, we developed a set of four MPRA libraries that cover more than 100 times the sequence space of the human genome (Fig. 1a and Methods ).
Discussion
Learning the rules by which DNA sequence determines where and when genes are expressed has proven surprisingly hard, despite the availability of full genome sequences of several mammals, extensive maps of genomic features 6, 11, 13 and genome-scale data about TF protein expression levels and TF DNA binding in vitro 2, 4, 5.
Methods
We designed a modified STARR-seq reporter construct pGL4.10-Sasaki-SS (a) based on an earlier published design 15 in the pGL4.10 backbone (Promega, E6651).
Data availability
All sequence data generated in this study are available under GEO accession GSE180158. All pretrained machine learning models are available at Zenodo with accession https://doi.org/10.5281/zenodo.5101420. Training, test and validation datasets for the CNN models are available at Zenodo with accession https://doi.org/10.5281/zenodo.5101420.
Acknowledgements
We thank M. Taipale, S. Wickström and M. Vartiainen for critical reading of the manuscript; A. M. Luoto, K. Jussila and Å. Kolterud for technical assistance; and L. Fei, J. Xia and N. Poddar for their help during the revision.
How does a gene regulatory network work?
During development each domain, where each domain represents a different spatial regions of the embryo, of gene expression will be under the control of different cis -regulatory modules. The design of regulatory modules help in producing feedback, feed forward, and cross-regulatory loops.
Which element is present on the same molecule of DNA as the gene they regulate?
To summarize, cis-regulatory elements are present on the same molecule of DNA as the gene they regulate whereas trans-regulatory elements can regulate genes distant from the gene from which they were transcribed.
Why is it important for an organism to have a gene?
The genome of an organism contains anywhere from a few hundred to thousands of different genes, all encoding a singular product or more. For numerous reasons, including organizational maintenance, energy conservation, and generating phenotypic variance, it is important that genes are only expressed when they are needed. The most efficient way for an organism to regulate gene expression is at the transcriptional level. CREs function to control transcription by acting nearby or within a gene. The most well characterized types of CREs are enhancers and promoters. Both of these sequence elements are structural regions of DNA that serve as transcriptional regulators.
How do enhancers affect transcription?
Multiple enhancers can act in a coordinated fashion to regulate transcription of one gene. A number of genome-wide sequencing projects have revealed that enhancers are often transcribed to long non-coding RNA (lncRNA) or enhancer RNA (eRNA), whose changes in levels frequently correlate with those of the target gene mRNA.
Why are cis regulatory modules difficult to identify?
Problems in identification arise because often scientists find themselves with a small set of known transcription factors, so it makes it harder to identify statistically significant clusters of transcription factor binding sites. Additionally, high costs limit the use of large whole genome tiling arrays.
What is a cis-regulatory module?
Cis -regulatory modules are non-random clusters at their specified target site that contain transcription factor binding sites. The original definition presented cis-regulatory modules as enhancers of cis-acting DNA, which increased the rate of transcription from a linked promoter.
Why are the architecture and the arrangement of the transcription factor binding sites critical?
The architecture and the arrangement of the transcription factor binding sites are critical because disruption of the arrangement could cancel out the function. Functional flexible cis -regulatory modules are called billboards. Their transcriptional output is the summation effect of the bound transcription factors.
Learning Objectives
Differences Between Prokaryotes and Eukaryotes: Mechanisms of Co-Regulation
- Often a set of proteins are needed together to respond to a certain stimulus or carry out a certain function (for example, many metabolic pathways). There are often mechanisms to co-regulate such genes such that they are all transcribed in response to the same stimulus. Both prokaryotic and eukaryotic cells have ways of co-regulating genes, but they use very different mechanisms t…
Overall Differences and Similarities
- If you understand the similarities and differences in eukaryotic and prokaryotic gene regulation, then you know which of the following process are exclusive to eukaryotes, which are exclusive to prokaryotes, which occur in both, and how each is accomplished: 1. coupled transcription and translation 2. 5′ cap and 3′ poly(A) tail 3. AUG as the translation initiation codon 4. regulation of …
Putting It All Together: Thelac Operon in E. Coli
- The lac operon is a good model gene for understanding gene regulation. You should use the information below to make sure you can apply all of the details of gene regulation described above to a specific gene model. E. coli lacoperon: dual positive and negative regulation The lac operon of E. coli has 3 structural genes required for metabolism of la...