A restriction enzyme, restriction endonuclease, or restrictase is an enzyme that cleaves DNA into fragments at or near specific recognition sites within molecules known as restriction sites. Restriction enzymes are one class of the broader endonuclease group of enzymes.
What function does a restriction enzyme serve?
Restriction enzymes, (aka endonucleases and exonuclease) are enzymatic proteins that bacteria express to defend against virus (phage) infections. Their function is to chop up the exogenous viral DNA or RNA before it can incorporate into the bacterial genome, hijack the cell, and start replicating more virus particles.
Why do we use restriction enzymes?
Restriction enzymes can be used to map DNA fragments or the entire genome, thus determining the specific order of the restriction enzyme sites in the genome. Restriction enzymes are also frequently used to verify the identity of a specific DNA fragment, based on the known restriction enzyme sites sequence that it contains. An extremely important use of restriction enzymes has been in the generation of recombinant DNA molecules.
Why is my restriction enzyme not cutting DNA?
Unexpected cleavage pattern—other
- The restriction enzyme tube or reaction buffer tube may be contaminated with a second enzyme. ...
- Another cause might be contamination of the DNA substrate. ...
- In rare cases, it may be possible that there are unexpected recognition sites in the substrate DNA. ...
- Finally, some restriction enzymes have degenerate recognition sites. ...
What do restriction enzymes do to the DNA?
Restriction enzymes dismantle foreign DNA by cutting it into fragments. This disassembling process is called restriction. Recombinant DNA technology relies on restriction enzymes to produce new combinations of genes. The cell protects its own DNA from disassembly by adding methyl groups in a process called modification.

What is restriction enzyme and its types?
Types of Restriction EnzymesType I. These restriction enzymes cut the DNA far from the recognition sequences. ... Type II. These enzymes cut at specific positions closer to or within the restriction sites. ... Type III. These are multi-functional proteins with two subunits- Res and Mod. ... In Gene Cloning.
Why is it called restriction enzyme?
Restriction enzymes were named for their ability to restrict, or limit, the number of strains of bacteriophage that can infect a bacterium.
What are the restriction enzymes with two examples?
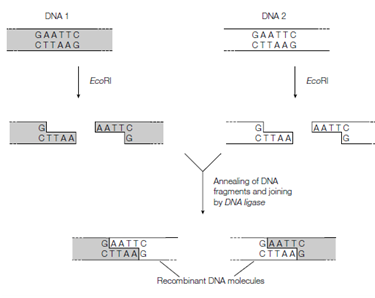
- EcoRI and smaI are the two examples of restriction enzymes. - The source of EcoRI restriction enzyme is E. coli RY 13 bacteria. EcoRI makes a staggered cut in DNA creating two sticky ends.
How do you write restriction enzymes?
A restriction enzyme is named with a prefix based on the first letter of the genus and first two letters of the species from which the enzyme was isolated. A fourth letter may be added to represent the species strain/serotype. Historically, the first three letters were italicized.
What is Type 4 restriction enzyme?
A fourth type (type IV) – methylation-dependent restriction enzymes (MDREs) – are REs that preferentially target modified DNA containing glycosylated bases, or methylated on adenine or cytosine residues, but lack a cognate MTase altogether.
Where are restriction enzymes found?
Restriction enzymes are found in bacteria (and other prokaryotes). They recognize and bind to specific sequences of DNA, called restriction sites.
What are the properties of restriction enzymes?
In general, restriction enzymes cleave double-stranded DNA. Each restriction enzyme recognizes specific DNA sequences, and cleavage can occur within the recognition sequence or some distance away, depending on the enzyme.
Who discovered restriction enzyme?
Werner ArberWerner Arber started this field of research in Geneva during the 1960's. He discovered restriction enzymes. Arber was studying an earlier known phenomenon, “host controlled restriction of bacteriophages”, and found that this process involved changes in the DNA of the virus.
How many restriction enzymes are there?
Approximately 3,000 restriction enzymes, recognizing over 230 different DNA sequences, have been discovered. They have been found mostly in bacteria, but have also been isolated from viruses, archaea and eukaryotes.
What is another name for restriction enzymes?
A restriction enzyme, restriction endonuclease, or restrictase is an enzyme that cleaves DNA into fragments at or near specific recognition sites within molecules known as restriction sites.
Why do we need restriction enzymes?
Restriction enzymes have proved to be invaluable for the physical mapping of DNA. They offer unparalleled opportunities for diagnosing DNA sequence content and are used in fields as disparate as criminal forensics and basic research.
What is restriction enzyme PDF?
A restriction enzyme is a site-specific endonucle- ase encoded by bacteria and archaea that recog- nizes a specific, short nucleotide sequence and. cuts the DNA only at that specific site, i.e., restriction site.
Why are restriction enzymes also called restriction endonucleases?
Introduction. Restriction enzymes are also called "molecular scissors" as they cleave DNA at or near specific recognition sequences known as restriction sites. These enzymes make one incision on each of the two strands of DNA and are also called restriction endonucleases.
Who discovered restriction enzyme?
Werner ArberWerner Arber started this field of research in Geneva during the 1960's. He discovered restriction enzymes. Arber was studying an earlier known phenomenon, “host controlled restriction of bacteriophages”, and found that this process involved changes in the DNA of the virus.
Why do restriction enzymes not cut their own DNA?
Restriction enzymes present in micro-organisms do not cut their own DNA because of the presence of the group that blocks digestion.
What is the origin of restriction endonucleases?
The term restriction enzyme originated from the studies of phage λ, a virus that infects bacteria, and the phenomenon of host-controlled restriction and modification of such bacterial phage or bacteriophage.
How do we Define a Restriction Enzyme?
A restriction enzyme is an enzyme produced by certain bacteria, which helps in the cutting or cleaving of the deoxyribonucleic acid (DNA) into smaller parts or fragments in any molecule. The difference between a restriction enzyme and any other endonucleases is that the restriction enzymes cleave at specific points known as restriction sites. A restriction enzyme is used as an important tool for genetic engineering.
What are restriction enzymes used for?
Restriction enzymes are widely used in the field of genetic engineering and bio-chemicals. These enzymes are used for cloning, especially type II of the restriction enzymes are used for cloning purposes. The modification function of the enzymes called methyltransferase or DNA methylation is used for genetic engineering.
What is a restriction fragment?
A restriction fragment refers to the fragment of the DNA that is produced after the cutting of the DNA strand and is caused by a restriction enzyme. Every restriction enzyme is extremely specific and recognizes a fort DNA sequence. Restriction enzymes are widely used in the field of genetic engineering and bio-chemicals.
What type of restriction enzyme cuts DNA away from the recognition sequence?
Type III Restriction Enzymes. Type III restriction enzymes are multifunctional proteins. This type of restriction enzyme cuts the DNA away from the recognition sequence. They have two subunits that carry the function of DNA methylation or modification and restriction digestion.
What type of enzymes are used to cleave DNA?
The type I restriction enzymes were the first restriction enzymes to be identified. These enzymes are characterized by their DNA cleavage sites. Type I enzymes cut DNA far away from the recognized sequence in the DNA molecule. They do not cause effective fragmentation of the DNA and hence, are of not much importance. Earlier, they were thought to be rare in nature, but continuous study and research proved that these type I enzymes are pretty common in nature. It is multifunctional as the type I restriction enzymes have three subunits that perform restriction digestion, recognition, and also modification of the DNA with the help of its cofactors like magnesium ions and ATP (adenosine triphosphate) that fulfill the catalyzing activity of the enzyme.
What are the three types of restriction enzymes?
Naturally occurring restriction enzymes list can be commonly divided into three major types, namely, Type I, Type II, and Type III. These are so grouped on the basis of their composition, nature of their target, cleavage position, and their enzyme cofactors (enzyme cofactors are chemical compounds that help enzymes in their catalyzing activities).
Why are restriction enzymes important?
A restriction enzyme is used as an important tool for genetic engineering. Bacteria use restriction enzymes to protect themselves from a dangerous virus called a bacteriophage, which translates to bacteria eater in literal terms. These attack bacteria and try to infect them by inserting their DNA in the cells of the bacteria.
How does restriction enzyme work?
Restriction enzyme function in the natural world is to defend bacteria against specific viruses called bacteriophages. These viruses attack bacteria by injecting viral RNA or DNA into a bacterial plasmid (small, purple ring in the below image) and replicating there . If viral RNA or DNA is detected within a prokaryote cell, that cell can often stop the replication process by slicing through the foreign genetic information. This renders it useless. At the same time, bacterial DNA is protected from the cutting action of its restriction endonucleases within its restriction sites. This mechanism adds methyl (H 3 C) groups to the cytosine and adenine of bacterial DNA without affecting the coded DNA sequence.
What are the different types of restriction enzymes?
Restriction Enzyme Groups. Natural restriction enzymes are arranged in five groups: type I, II, III, IV, and V. Type I REs, the first to be discovered, cut DNA sequences far from the recognition sites and require ATP to recognize, modify and/or digest asymmetrical sections.
Why are artificial restriction enzymes used in CRISPR?
Furthermore, commercially-available natural restriction enzymes are limited in number, and these fragment DNA into very short sections; it is rare that a smaller laboratory has access to the right enzymes. This is because different restriction enzymes are required to cleave the many separate areas of DNA that make up the code for a single gene.
Why are restriction endonucleases rarely used in genetic engineering?
Type III restriction endonucleases are rarely used in genetic engineering as they cut DNA sequences well outside of the recognition sequence and need to detect two separate sequences to achieve this. This means they are not always able to provide complete restriction enzyme digestion.
What type of enzyme is used for DNA mapping?
Type II restriction enzymes (REs) are of particular importance in the fields of molecular cloning, gene sequencing, and DNA mapping as this group can cut DNA very close to specific recognition sites and does not require energy in the form of ATP. Restriction enzymes – molecular scissors.
How does DNA ligase work?
DNA ligase then recombines the DNA by constructing a mirror copy of the bacterial sequence. In short, a restriction enzyme cleaves the foreign DNA and DNA ligase repairs the break to bring it back to its original form. Since the discovery of genes, ways to manipulate them have been heavily researched.
How many restriction enzymes are there in plasmids?
To date, approximately 3500 restriction enzymes have been isolated from bacterial plasmids. Each enzyme recognizes a specific sequence of viral genetic code and will try to separate the new, mutated DNA strand close to or further away from the recognition site.
What is restriction enzyme?
Restriction enzymes are enzymes that cut DNA into fragments based upon recognizing a specific sequence of nucleotides. Restriction enzymes are also known as restriction endonucleases. Regina Bailey is a board-certified registered nurse, science writer and educator. Her work has been featured in "Kaplan AP Biology" and "The Internet for Cellular ...
Why are restriction enzymes important?
Restriction enzymes have been used to help produce vaccines, pharmaceutical products, insect resistant crops, and a host of other products.
What Is DNA Ligase?
The sticky ends of the fragments produced by restriction enzymes are useful in a laboratory setting. They can be used to join DNA fragments from both different sources and different organisms. The fragments are held together by hydrogen bonds. From a chemical perspective, hydrogen bonds are weak attractions and are not permanent. Using another type of enzyme however, the bonds can be made permanent.
What sequences do restriction enzymes recognize?
For example, a restriction enzyme may recognize a specific sequence of guanine, adenine, adenine, thymine, thymine, cytosine. When this sequence is present, the enzyme can make staggered cuts in the sugar-phosphate backbone in the sequence. But if restriction enzymes cut based on a certain sequence, how do cells like bacteria protect their own DNA ...
What happens when restriction enzyme cuts DNA?
Since the DNA is cut on both strands, there will be complementary ends that can hydrogen bond to one another. These ends are often called "sticky ends."
What enzymes dismantle DNA?
Restriction enzymes dismantle foreign DNA by cutting it into fragments. This disassembling process is called restriction.
What is the recognition sequence of an enzyme?
Each enzyme has what is known as a recognition sequence or site. A recognition sequence is typically a specific, short nucleotide sequence in DNA. The enzymes cut at certain points within the recognized sequence. For example, a restriction enzyme may recognize a specific sequence of guanine, adenine, adenine, thymine, thymine, cytosine.
Definition
Restriction Enzyme Function
- Restriction enzyme function in the natural world is to defend bacteria against specific viruses called bacteriophages. These viruses attack bacteria by injecting viral RNA or DNA into a bacterial plasmid (small, purple ring in the below image) and replicating there. If viral RNA or DNA is detected within a prokaryote cell, that cell can often stop the replication process by slicing throu…
Restriction Enzyme Groups
- Natural restriction enzymes are arranged in five groups: type I, II, III, IV, and V. Type I REs, the first to be discovered, cut DNA sequences far from the recognition sites and require ATP to recognize, modify and/or digest asymmetrical sections. The distance from the recognition site makes type I restriction endonucleases less helpful in the field of genetic engineering. Type II REs recognize a…
Traditional DNA Cloning Uses
- Traditional DNA cloning using restrictive endonucleases has multiple uses. Expressed recombinant DNA(DNA sequences that code for protein synthesis), when inserted into the genetic information of bacteria, stimulate bacteria to produce the target protein. This is the method whereby genetic engineers in pharmaceutical companies manufacture human insulin, human al…